Scientists demonstrate modulation of gene expression by protein coding regions
A research team at the Stowers Institute has discovered how the expression of one of the Hox master control genes is regulated in a specific segment of the developing brain. The findings provide important insight into how and where the brain develops some of its unique and important structures.
The findings were posted to the online Early Edition of the Proceedings of the National Academy of Science today.
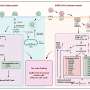
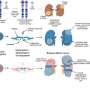
The team led by Robb Krumlauf and Leanne Wiedemann set out to understand the "instruction manual" for a Hox gene that tells the early brain which genes to turn on and in what order, to specify critical regions of the adult brain. Their studies discovered how expression of the key regulatory protein, encoded by the Hoxa2 gene, is controlled. Surprisingly, the DNA sequence that contains the instructions about when and where to express Hoxa2 in a segment of the developing brain overlaps with sequences that code for amino acids of the Hoxa2 protein.
"In the mammalian genome, sequences that encode proteins and those that control gene expression are usually separate from each other," explained Robb Krumlauf, Ph.D., Scientific Director. "Most approaches to the identification of DNA elements that control gene expression utilize methods that exclude protein coding domains. Our group has now discovered that protein coding regions can also play a role in modulating gene expression. This work has important implications for identifying the regulatory logic contained in mammalian genomes."
"Our findings provide important insight into the regulation of the formation of the anterior hindbrain," said Leanne Wiedemann, Ph.D., a co-investigator in the Krumlauf Lab and senior author on the publication. "Additionally, because we now understand that regulatory input from coding regions needs to be considered, our findings have broader implications in helping to design tests and interpret data from large-scale analyses of gene regulation."
Expanding on this work, their lab will continue to dissect the regulatory networks and integrate the genes that play a role in hindbrain development using evolutionary comparisons, bioinformatics approaches, and experimental analyses.
Source: Stowers Institute for Medical Research