Analysis of sperm differentiation reveals new mode of proteasome regulation
(Medical Xpress) -- Early in development, cells undergo a controlled demolition that helps to shape their raw, pliable material into the specialized forms they must have to do their jobs as adults. The process by which this occurs is also crucial later in the cell’s life, to take out potentially dangerous trash that routinely accumulates as it ages. New work from Rockefeller University has now provided a detailed genetic and biochemical understanding of how one protein helps modulate this sculptor and janitor of the cell, known as the proteasome. The findings improve our knowledge of how cells specialize, and could also lead to new tools for fighting cancer and degenerative disorders.
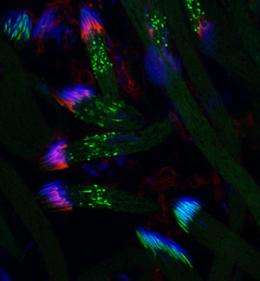
A good place to study the proteasome in action is in the developing sperm cell, which prunes away much of its cell mass to cut a sleek and efficient swimmer. “It’s got to be a light, efficient DNA delivery vehicle,” says Hermann Steller, head of the Strang Laboratory of Apoptosis and Cancer Biology. Through a genetic screen of sterile, male flies, the team isolated a protein, called Nutcracker, without which sperm cell remodeling does not occur, causing sterility. Nutcracker is part of the larger SCF complex, which helps regulate the destruction of proteins involved in cell cycle control, among other things.
Maya Bader, a former doctoral student in Steller’s lab, undertook the arduous dissection of 10,000 fruit fly testes to screen for proteins that were interacting with Nutcracker. Using mass spectrometry, she identified DmPI31, a molecule originally described as an inhibitor of proteasome activity in mammalian cells about 10 years ago. But the researchers were in for a surprise: probing the function of DmPI31 with genetic and biochemical experiments, they found that it did not inhibit but rather enhanced proteasome activity, and that its absence was lethal, according to experiments to be published April 29 in Cell.
Digging deeper into the interaction of Nutcracker and DmPI31, Bader and colleagues built on research she had published last year detailing which part of Nutcracker facilitated its interaction with the SCF complex. Creating mutants of Nutcracker and observing the effects on their interaction with DmPI31, the team found that a part of Nutcracker, called the FP domain, was the key player. They found that mutating the FP domain of Nutcracker in flies prevented the interaction with DmIP31, and the result was sterility. On the other hand, creating flies that generated extra DmPI31 partially reversed this effect. They concluded that Nutcracker and DmPI31 must interact via the FP domain in order for Nutcracker to regulate DmPI31 and to achieve the normal “controlled demolition” that creates effective sperm cells.
“Until recently, much of the work on proteasomes has been carried out using in vitro systems and cultured cells. By studying proteasome function in developing animals, we were able to unveil a new mode of proteasome regulation that is important for normal development,” Bader says. “The fact that proteasomes are themselves regulated demonstrates that the cells can change 'proteolytic states' to facilitate normal developmental processes, a change which also occurs in the progression of protein homeostasis diseases."
Looking beyond fly sperm cells, the researchers, in collaboration with David Smith in Alfred Goldberg’s laboratory at Harvard University, also studied the activity of DmPI31 in isolated mammalian proteasomes. They found that its effect on the dominant form of the proteasome in living cells was to activate it, despite its original description as an inhibitor. In fact, in flies that have problems that result from reduced proteasome function, increasing the expression of DmPI31 corrected the problem.
In flies in which the gene for DmPI31 was mutated to prevent the expression of the protein, protein degradation was compromised and the larvae died at the early pupal transition, a developmental stage where many cell cycle related mutants are lethal. In sum, the experiments show that DmPI31, which functions in both mammalian and fly cells, is a crucial activator of the proteasome, which is a dynamically regulated system that performs controlled demolition, remodeling cells during development. It’s also critical in routine cell maintenance, “taking out the trash” that accumulates in cells later in life.
“The discovery of a novel way to regulate the proteasome’s activity sheds new light on how controlled protein destruction helps shape cell specialization during development,” said Marion Zatz, who oversees Steller’s and other programmed cell death grants at the National Institutes of Health. “This study adds a new dimension to what we know about how organisms develop, and could lead to insights on how misregulated protein degradation contributes to neurodegenerative diseases like Alzheimer’s.”
Steller’s and Bader’s Harvard collaborator, Goldberg, pioneered research that led to the development of a proteasome inhibitor, marketed as Velcade, that is used to treat multiple myeloma, so the medical relevance of manipulating proteasome function has already been established. The new work could eventually lead to new drugs that target proteasomes for therapeutic benefits, Steller says.
“Controlled proteolysis is essential for many cell biological functions,” says Steller, who is also a Howard Hughes Medical Institute investigator. “There had been the impression that the proteasome is just a brute ‘shredder,’ but it doesn’t run at full steam all the time. It’s modulated, and these findings give us new ideas for designing small molecules that regulate proteasome activity.”
More information: Cell 145: 371–382 (April 29, 2011) A Conserved F Box Regulatory Complex Controls Proteasome Activity in Drosophila, by Maya Bader, Sigi Benjamin, Orly L. Wapinski, David M. Smith, Alfred L. Goldberg and Hermann Steller. www.cell.com/abstract/S0092-8674(11)00298-4