Researchers discover new bone deformity gene
(Medical Xpress) -- The Human Genetics team at The University of Queensland Diamantina Institute have successfully used a new gene-mapping approach for patients affected by severe skeletal abnormalities.
Skeletal dysplasias are a group of diseases that cause abnormalities in the skeleton's growth and function. This can lead to problems such as abnormal height and/or limb length, difficulty with reproduction and decreased life span. Families affected by skeletal dysplasias are usually very small in number, which can make it difficult to find the disease-causing gene for that family.
Associate Professor Andreas Zankl, a clinical geneticist from The University of Queensland Centre for Clinical Research, developed a Bone Dysplasia registry for patients and their families – the first of its kind in Australia. Through the registry, the UQDI team of researchers met a family with two young daughters affected by a severe form of dwarfism.
The team used next-generation sequencing to simultaneously study the four immediate family members and compare their exomes – the coding section of the genes – to each other and against the reference sequence from the international Human Genome Project.
They were able to discover which gene within the family caused the abnormality. Impressively, the mapping process took only a few weeks. The UQDI researchers then successfully determined how the genetic abnormality caused the skeletal disease.
In the past, researchers could only sequence and compare a few genes at a time, which was expensive and time-consuming. For example, UQDI researchers had spent a decade finding the responsible gene for another type of skeletal dysplasia, fibrodysplasia ossificans progressiva.
In contrast, next-generation sequencing technology can provide more rapid results for mapping genes in these particular types of diseases. However, despite this breakthrough in progress, Associate Professor Emma Duncan said it was still an intensive process.
"Typically, we all have a number of small genetic differences – we find approximately 20,000 on average just in our coding regions when compared with the Human Genome sequence – so it's still a very involved process to work out which one is the disease-causing mutation," she said.
"For this family, it's been a huge relief to find out why their little girls have this devastating skeletal disorder, and understanding the genetics has helped them in their planning for any future pregnancies," said Professor Matthew Brown.
With the success of their next-generation sequencing approach, the team have also researched another skeletal dysplasia case which involved five unrelated individuals, comparing their exomes with each other and with the Human Genome Project.
By examining just this small number of affected people, the responsible gene has been identified. UQDI researchers will continue to map unknown genes for skeletal dysplasias and for other likely single-gene inherited diseases.
The paper has been published in the Public Library of Science (PLoS) Genetics.
More information: Glazov EA, Zankl A, Donskoi M, Kenna TJ, Thomas GP, et al. (2011) Whole-Exome Re-Sequencing in a Family Quartet Identifies POP1 Mutations As the Cause of a Novel Skeletal Dysplasia. PLoS Genet 7(3): e1002027. doi:10.1371/journal.pgen.1002027
Abstract
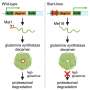
Recent advances in DNA sequencing have enabled mapping of genes for monogenic traits in families with small pedigrees and even in unrelated cases. We report the identification of disease-causing mutations in a rare, severe, skeletal dysplasia, studying a family of two healthy unrelated parents and two affected children using whole-exome sequencing. The two affected daughters have clinical and radiographic features suggestive of anauxetic dysplasia (OMIM 607095), a rare form of dwarfism caused by mutations of RMRP. However, mutations of RMRP were excluded in this family by direct sequencing. Our studies identified two novel compound heterozygous loss-of-function mutations in POP1, which encodes a core component of the RNase mitochondrial RNA processing (RNase MRP) complex that directly interacts with the RMRP RNA domains that are affected in anauxetic dysplasia. We demonstrate that these mutations impair the integrity and activity of this complex and that they impair cell proliferation, providing likely molecular and cellular mechanisms by which POP1 mutations cause this severe skeletal dysplasia.