New automated imaging to greatly speed whole-brain mapping efforts
A new technology developed by neuroscientists at Cold Spring Harbor Laboratory (CSHL) transforms the way highly detailed anatomical images can be made of whole brains. Until now, means of obtaining such images – used in cutting-edge projects to map the mammalian brain -- have been painstakingly slow and available only to a handful of highly specialized research teams.
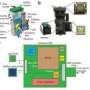
By automating and standardizing the process in which brain samples are divided into sections and then imaged sequentially at precise spatial orientations in two-photon microscopes, the team, led by Assoc. Prof. Pavel Osten and consisting of scientists from his CSHL lab and the Massachusetts Institute of Technology, has opened the door to making whole-brain mapping routine.
Specifically, says Osten, "the new technology should greatly facilitate the systematic study of neuroanatomy in mouse models of human brain disorders such as schizophrenia and autism."
The new technology, developed in concert with TissueVision of Cambridge, Mass. and reported on in a paper appearing online Jan. 15 in Nature Methods, is called Serial Two-Photon Tomography, or STP tomography. Tomography refers to any process (including the familiar CAT and PET scans used in medical diagnostics) that images an object section by section, by shooting penetrating waves through it. Computers powered by mathematical formulae reassemble the results to produce a three-dimensional rendering. Two-photon imaging is a type used in biology laboratories, particularly in conjunction with fluorescent biomarkers, which can be mobilized to illuminate specific cell types or other anatomical features. The two-photon method allows deeper optical penetration into the tissue being sampled than conventional confocal microscopy.
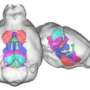
As Osten explains, STP tomography achieves high-throughput fluorescence imaging of whole mouse brains via robotic integration of the two fundamental steps -- tissue sectioning and fluorescence imaging. In their paper, his team reports on the results of several mouse-brain imaging experiments, which indicate the uses and sensitivity of the new tool. They conclude that it is sufficiently mature to be used in whole-brain mapping efforts such as the ongoing Allen Mouse Brain Atlas project.
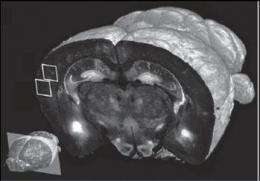
One set of experiments tested the technology at different levels of resolution. At 10x magnification of brain tissue samples, they performed fast imaging "at a resolution sufficient to visualize the distribution and morphology of green-fluorescent protein-labeled neurons, including their dendrites and axons," Osten reports.
A full set of data, including final images, could be obtained by the team in 6.5 to 8.5 hours per brain, depending on the resolution. These sets each were comprised of 260 top-to-bottom, or coronal, slices of mouse brain tissue, which were assembled by computer into three-dimensional renderings themselves capable of a wide range of "warping," i.e., artificial manipulation, to reveal hidden structures and features.
"The technology is a practical one that can be used for scanning at various levels of resolution, ranging from 1 to 2 microns to less than a micron," Osten says. Scans at the highest resolution level take about 24 hours to collect. This makes possible an impressive saving of time, Osten says, compared to methods that are now in use. Using these, it would take an experienced technician about a week to collect a set of whole-brain images at high resolution, he noted.
"What is most exciting about this tool is its application in the study of mouse models of human illness, which we are already doing in my lab," Osten says. "We are focusing on making comparisons between different mouse models of schizophrenia and autism. Many susceptibility genes have been identified in both disorders – one recent estimate by Dr. Mike Wigler's team here at CSHL put the figure at over 250 for autism spectrum disorders, for instance. Dr. Alea Mills at CSHL has published a mouse model of one genetic aberration in autism – a region on chromosome 16 – and soon we will have tens of models, each showing a different aberration.
"We will want to compare these mice, and that is essentially why we designed STP tomography – to automate and standardize the process of collecting whole-brain images in which different cell-types or circuit tracings have been performed. This makes possible comparisons across different mouse models in an unbiased fashion."
More information: "Serial two-photon tomography: an automated method for mouse brain imaging," Nature Methods on January 15, 2012.