Parallel evolution - cystic fibrosis
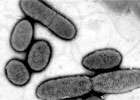
(Medical Xpress) -- Researchers at the University of Liverpool are examining the evolution of Pseudomonas aeruginosa over time in the lungs of ten people with cystic fibrosis to understand why it persists in these patients and why its response to antibiotics is so varied.
There's no getting away from bacteria: we breathe in thousands of them every day. Most of them don't do us much harm, but some - such as certain strains of Streptococcus, Staphylococcus and Pseudomonas - have the potential to cause severe infections like pneumonia and pleurisy if they get into our lungs.
Fortunately, most healthy people have a series of mechanical and biological barriers - including the mucus and cilia in our respiratory tract (our mucociliary escalator) - that prevent bacteria we inhale getting into our lungs.
For people with cystic fibrosis, however, it's a different story. A genetic mutation means they produce a thick, sticky secretion that lines their respiratory tract, severely impairing their mucociliary escalator. Consequently, harmful bacteria can get into the lungs and stay there, safely cocooned inside protective biofilms or sacs, which they make by secreting polysaccharides.
"Once these bacteria colonise the lung of someone with cystic fibrosis, it's impossible to get rid of them completely with antibiotics," says Professor Craig Winstanley at the Institute of Infection and Global Health at the University of Liverpool. The bacteria remain inside the person's lungs for the rest of his or her life, causing the severe lung infections that eventually kill people with cystic fibrosis.
Until around 15 years ago, clinicians believed that these harmful strains weren't transmissible between people with cystic fibrosis. In 1996, however, Craig and colleagues identified a strain of Pseudomonas aeruginosa that not only caused particularly severe infections in people who caught it but also could spread from patient to patient.
"Children with cystic fibrosis used to go on summer camps together, but once people realised they could pass nasty strains of lung infections on to each other, of course that had to stop," he says. "In our clinic here, we segregate cystic fibrosis patients - they come in for treatment on different days - so that they can't pass on virulent strains to each other."
Because this particular strain of P. aeruginosa was first identified in Liverpool, Craig and colleagues named it the 'Liverpool epidemic strain' (LES). They developed a rapid diagnostic test to identify LES in spit taken from people with cystic fibrosis, based on the DNA sequence in specific regions of its genome (a procedure known as a molecular typing assay).
Yet although they can now rapidly identify the LES strain, clinicians have remained baffled by the fact that antibiotics targeting the strain have dramatically different effects in different patients - even between people with the same infection. "It's hit and miss whether a particular antibiotic will work," says Craig. "I'm afraid we're still in the Dark Ages when it comes to treating infections in these patients.
"Sometimes a patient gets better, sometimes not, and we don't know why. We don't even know what antibiotics are doing to infections, biologically. They're certainly not removing them."
What if
To try to understand this conundrum, Craig asked himself the all-important question that so often drives scientific advances: what if? What if antibiotics have such varied effects on LES infections because of evolutionary forces acting on the bacteria in the lung?
"The prevailing idea is that infections in CF patients are caused by a single, genetically uniform bacterium," he says. "Microbiologists take a sputum sample from the patient and typically isolate one P. aeruginosa bacterium in the sample. They then test that single isolate for which antibiotics might be effective against it and send the answer to the clinicians."
This approach is perfectly valid for an infection like a burn, which heals (and kills off the infecting bacteria) rapidly in healthy people, but bacteria that get into the lungs of people with cystic fibrosis live there for years. What if, during that time, the original infecting bacterium began to mutate, producing clones that were genetically different from the 'parent'? Could there be genetically different versions of the LES strain living within people with cystic fibrosis - accounting for the varying effects of antibiotics aimed against the original 'parent' bacterium?
To test that hypothesis, Craig decided to look at several people with cystic fibrosis with the same infection over a period of time. In 2009, he and his colleagues took sputum samples at monthly intervals from ten people with cystic fibrosis infected with the same LES strain of P. aeruginosa.
They isolated and grew clones of not one but 40 whole bacteria - all representatives of LES - from every sputum sample, resulting in a total of 1720 isolates, or clones. They then characterised each of these 1720 clones for 15 traits or measurable behaviours (phenotypes).
These included what each clone actually looked like on a petri dish (its colour and morphology), whether it could grow in certain sorts of media, what (if any) antibiotics it was resistant to, what toxins it secreted and whether it had lost the ability to use glucose as its only source of carbon. They also did some genetic tests, helped by Dr Steve Paterson at the University's Centre for Genomic Research, to look for specific chunks of DNA and bits of the chromosome that were unstable.
Using these 15 traits or phenotypes, they built up a profile of each of these 1720 clones and compared them to see whether they were identical or could be classed as different types of the same strain.
The results, published in the American Journal of Respiratory and Critical Care Medicine in 2011, revealed a startling amount of diversity across these 15 traits. "We found hundreds of distinct subtypes of the same strain. A lot more than probably people had imagined," says Craig. Most surprisingly of all, most of that phenotypic or behavioural diversity occurred in individual patients rather than being due to differences between patients.
The bacteria had undergone rapid genetic mutations (over a period of months) in the lung that enabled them to change their behaviour. This explains the varied response to antibiotics and other treatments seen to date, and why it is so difficult to decide which antibiotic to use for a successful outcome.
"In fact," says Craig, "the current tests are completely meaningless, although they're widely used. And everyone's starting to recognise this, but nobody knows what the alternatives are. The thing that doesn't help is not recognising that this happens."
"Taking 40 individual little colonies per sample and taking lots of samples over time is what really revealed that surprising diversity. It has given an entirely novel outlook on these infections," says Steve.
Zooming in
Because these changes in the bacteria are clearly very important, enabling them to survive and thrive in people with cystic fibrosis, it's crucial to understand how, when and why they occur.
To get a better handle on those mutations - and the biological changes they generate - Steve and Craig now have a three-year Wellcome Trust project grant to zoom in more precisely on the isolates collected in 2009. They will be collaborating with Dr Mike Brockhurst at the Institute of Integrative Biology, who will be applying techniques from population genetics and evolutionary biology to the dataset.
Steve, Mike and colleagues will be using next-generation high-throughput DNA sequencing technology to sequence the entire chromosome of representative batches of isolates from Craig's ten patients sampled in 2009.
The Centre for Genomic Research at Liverpool boasts a Life Sciences SoLiD 5500 XL machine - the first of its kind in the UK. "It works 250 000 times faster than the technology used to sequence the human genome ten years ago," says Steve. "It can read 30 billion letters of DNA sequence per day, compared to four billion using current machines. That's the equivalent of about 100 human genomes in five days. So I'll be able to sequence around 100 bacterial genomes in a week.
The technology will allow them to investigate the mutations in the LES bacteria at a much finer resolution than anyone has ever been able to do before. "We can sequence multiple bacteria and compare things down to individual changes in the base pairs in the gene sequences."
Predicting evolution
An important and unique aspect of the project is that it is the first time the evolution of LES has been followed in real, living people, rather than in the laboratory, in test tubes and artificial systems. What particularly excites Steve is the fact that because each of the ten people from which the samples were collected had the same strain of LES, its evolution has been set up in parallel, but independently.
Because bacteria reproduce rapidly (every half an hour or so), Steve will be able to see mutations occurring within the space of months or weeks. Following the mutations as they occur should help shed light on how the bacteria adapts so efficiently to people with cystic fibrosis and what changes in the bacterial populations account for its success.
"These patients are being hit by a lot of antibiotic drugs as part of their standard therapy, so the bugs evolve a response to that. We'll try and find what mutations allow these bugs to become antibiotic resistant or what changes allow them to secrete toxins into the lungs," says Steve.
They will also try to establish the changes or fluctuations in the make-up of the bacterial population that might make the infection more virulent or trigger an exacerbation in people. A lot of damage is done to the lung during these exacerbations, and patients have to be hospitalised and given intravenous antibiotics.
The length of the infection - and its course in ten independent environments, or patients - will also answer an important evolutionary question: is evolution repeatable? Does the strain evolve the same way in different people, or will the researchers find ten completely separate outcomes?
If certain types of mutations are repeatable - if we can predict which way the bacteria is going to jump, and if there are changes that all the bacteria undergo - that might help us identify weak points in the bacteria, which could be targets for new types of interventions or drugs," says Steve.
"This project is trying to address a huge gap in our understanding of what the bacteria are actually doing inside the patients," says Craig, "because the methods that are used at the moment don't actually get rid of them. Hopefully, we'll be able to pass this back to the clinicians later on in the project, and start improving outcomes for patients living with cystic fibrosis."
More information: Winstanley C. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am J Respir Crit Care Med 2011;183(12):1674-9.
Fleming S. Relating introspective accuracy to individual differences in brain structure. Science 2010;329(5998):1541-3.