Taking a hit or two
Cancer only arises if two or more genes are mutated. Learning which combinations of mutations cause cancer represents an extremely laborious endeavor. In the current issue of the journal Nature Methods Robert Eferl and colleagues announce a novel mouse model which is set to make this work much easier.
Despite a huge amount of research effort, the molecular mechanisms that underlie the transition from a "normal" cell to a cancerous cell are only poorly understood. After the discovery of the first cancer-causing genes or oncogenes and the finding that they are mutated forms of normal cellular genes, it was widely believed that a single mutation was enough to cause cancer. Subsequent research, however, has revealed that most cancers only develop as a result of several mutations. A bewildering variety of combinations of mutations have been shown to have the potential to give rise to cancer. Finding out which combinations are dangerous has to date been largely a matter of trial and error but this should change with the development of a tool to identify mutations that really do collaborate to cause cancer. Robert Eferl and colleagues announce the new "Multi-Hit" mouse in the current issue of the journal Nature Methods. The work is one result of a longstanding collaboration between many institutions in the Vienna area coordinated by Mathias Müller of the University of Veterinary Medicine, Vienna.
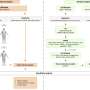
It is now generally accepted that cancer only arises if two or more genes are mutated. To date, learning which combinations of mutations cause cancer has represented an extremely laborious endeavour but the development of the "Multi-Hit" mouse looks set to change this. The group of Robert Eferl at the Ludwig Boltzmann Institute for Cancer Research, Vienna has taken advantage of the Cre-recombinase system to generate random combinations of correctly and incorrectly oriented oncogenes (or tumour suppressor genes, genes whose inactivation may contribute to the development of cancer) and investigated which of the combinations caused tumours.
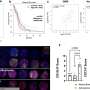
They tested their system on the well-known Ras protein, which has been shown to be mutated in many different cancers. Ras mutations were thought to cause cancer only if the so-called Raf gene was also mutated but it has more recently been proposed that changes in other genes, such as those encoding the RALGEF (Ral guanine nucleotide exchange factor), MAPK (mitogen-activated protein kinase) or PI3K (phosphatitylinositol-3-kinase) proteins, may also combine with mutated Ras proteins to cause tumour development. The researchers found that mutations in Ras alone did not cause tumours to develop, while following activation of the Cre recombinase (and thus random activation – by flipping – of the genes under study) all mice developed cancer.
Examination of the tumours showed that in most of them all three genes had been activated, although activation of the P13K gene alone (and in very rare cases of one of the other two genes alone) could also give rise to cancer. In other words, the most rapidly proliferating, and thus most life-threatening, tumours were associated with activation of all three of the genes investigated. This indicates that all the genes are somehow contributing to the development of cancer, which means that drugs targeting any or all of them could play a part in treatment.
Eferl, now at the Institute for Cancer Research & Comprehensive Cancer Center of the Medical University of Vienna, is naturally excited by the results. "Our work on Ras has given important clues to possible therapeutic strategies. But this was really only a proof-of principle. More importantly, the results show that our Multi-Hit mouse can indeed be used to study interactions between gene mutations. This should make it much easier for us to understand how cancer arises and what we can do to treat it."
More information: The paper "A mouse model to identify cooperating signalling pathways in cancer" by Monica Musteanu, Leander Blaas, Rainer Zenz, Jasmin Svinka, Thomas Hoffmann, Beatrice Grabner, Daniel Schramek, Hans-Peter Kantner, Mathias Müller, Thomas Kolbe, Thomas Rülicke, Richard Moriggl, Lukas Kenner, Dagmar Stoiber, Josef Penninger, Helmut Popper, Emilio Casanova and Robert Eferl is published in the current issue of Nature Methods ( DOI: 10.1038/nmeth.2130 ).