Researchers use supercomputer simulations to understand how some carcinogens evade removal
A person doesn't have to go far to find a polycyclic aromatic hydrocarbon (PAH). These carcinogen precursors are inhaled through automobiles exhaust during the morning commute, are present in a drag of cigarette smoke, and are part of any barbequed meal.
Once ingested or inhaled, these big, bulky multi-ringed molecules are converted into reactive carcinogenic compounds that can bind to DNA, sometimes literally bending the double helix out of its normal shape, to form areas of damage called lesions. The damaged DNA can create errors in the genetic code during replication, which may cause cancer-initiating mutations.
It is the job of the nuclear excision repair (NER) system to repair damage caused by PAH lesions by removing the segment of DNA where the lesion is bound and patching up the resulting gap. But some lesions are especially resistant to this repair machinery, making them much more likely to cause mutations than lesions that are promptly repaired.
A research team at New York University (NYU) has gained new insight on the ability of certain PAH-derived lesions to evade the DNA repair machinery. They found that some lesions stabilize the DNA they damage, making it difficult for a certain repair protein to mark the lesion for repair. Their research was published earlier this year in the February 2012 issue of Biochemistry. More recent articles about NER of DNA lesions from the same group appeared in Nucleic Acids Research in July and August.
"Some lesions cause DNA to be locally destabilized, but there are lesions that actually stabilize the DNA so that the two strands come apart with great difficulty," said Suse Broyde, a biology professor at NYU. "Sometimes they're even more stable than undamaged DNA."
The stability of the DNA double helix is a key feature that determines whether DNA is flagged for repair in the first place by a protein called XPC. The protein patrols the genome looking for weakened areas. When it finds one, it slips a structure called a beta-hairpin between the strands, marking the DNA for NER. But if a lesion makes DNA more stable, the strands become more difficult to separate and the beta hairpin can't signal for repair.
How can molecules as large and disruptive as a PAH stabilize DNA in the first place?
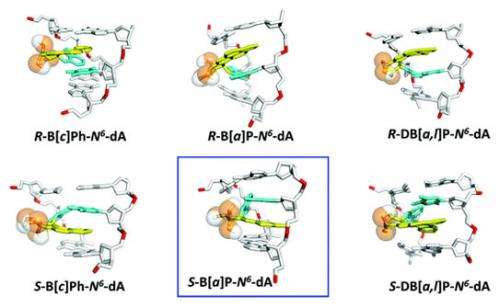
![Top: Models showing different lesions wedged between DNA bases. Lesions that are more easily repaired, like the one marked in the image, are more likely to be removed before they can create mutations in replicated DNA. Bottom:The stacking interactions between lesions and adjacent DNA base pairs stabilize DNA. Dibenzo [a,l] pyrene derived lesions (featured on the top and bottom right) have a great number of interactions, making them especially difficult to remove. Credit: New York University Cancer bound](https://scx1.b-cdn.net/csz/news/800a/2012/cancerbound.jpg)
"If you remember from chemistry, every kind of molecule interacts with other molecules through so-called Van der Waals interactions," said Nicholas Geacintov, a chemistry professor at NYU. "The DNA and the carcinogen bound to it also have the same kind of interactions," he said, specifically referring to the PAH-derived lesions that intercalate, or wedge themselves, between DNA base pairs.
The role of Van der Waals forces, which Broyde calls "stacking interactions" for these systems, was made clear through a series of computer simulations performed, analyzed, interpreted and visualized by Yuqin Cai, a post-doctoral senior research scientist in Broyde's lab. "The computer simulations revealed the structural, energetic and dynamic properties of the DNA containing the PAH-derived lesions, " said Broyde, who specializes in providing a mechanistic understanding of complex biological processes using molecular dynamics simulations and other front-line computational approaches.
"You can make movies of the dynamic trajectory which allow you to see the real mobility of the entire system," Broyde said. "You can watch the DNA flexing and the backbone moving dynamically and the carcinogen moving in and out. It's not rigid—you can see its aliveness."
Broyde and her team's simulations revealed that of the six different lesions examined (three chemicals with two different geometric configurations each), those caused by dibenzo[a,l]pyrene, the most tumorigenic PAH investigated, were the most resistant to repair. The five-ringed structure of the carcinogen provided ample stacking opportunities, which stabilized the DNA much better than the four and three-ringed structures of the other PAHs that were examined.
Knowing which lesions are the most repair resistant could play an important role in preventative medicine, said Broyde, as individuals harboring them could be counseled to avoid further exposure, particularly in the case of smokers.
The atomic-level visualizations Broyde's lab simulated explained data gathered from experiments that are carried out in Geacintov's lab, including synthesis of the damaged DNA, measurements of its stability, and investigations of the relative NER susceptibility of lesion-containing DNA in human cells.
"Without experiments they wouldn't have anything to model and without modeling it would be very hard for us to understand what we are measuring," Geacintov said.
Some of the most important results came from DNA melting experiments, where a specific sequence of double-stranded DNA was synthesized with a known lesion and then heated up until the strands separated. The more stable the DNA, the higher the temperature at which it melted. The most stabilizing lesion melted nearly 10 degrees higher than the melting temperature of DNA with no lesions; this lesion was the most repair-resistant in experiments and also most stabilizing according to molecular simulations.
To compute the raw data (coordinates of structures as a function of time) for the analyses and visualizations, the Broyde lab used the Longhorn, Lonestar and Ranger high-performance computing resources at the Texas Advanced Computing Center as well as other systems in the Extreme Science and Engineering Discovery Environment (XSEDE).
Uncovering the stabilizing properties that allow some lesions to evade NER and initiate cancer could help develop better chemotherapeutic drugs to fight the disease, according to Broyde and Geacintov.
For example, the widely used chemotherapy drug, Cisplatin, attacks the DNA of cancerous cells, interrupting unregulated replication. However, the cell's own NER machinery can combat the drug by removing it and repairing the genome, just as in healthy non-cancerous cells.
"One direction in drug design is to find pharmaceuticals that still inhibit replication but that are less susceptible to NER," Broyde said. "Understanding the mechanism of NER will be valuable in designing the next generation of chemotherapeutic agents. These would be more effective if they were more resistant to NER."