Neuroblastoma patients with ARID1A and ARID1B mutations have more aggressive disease
In a genome sequencing study of 74 neuroblastoma tumors in children, scientists at the Johns Hopkins Kimmel Cancer Center and the Children's Hospital of Philadelphia (CHOP) found that patients with changes in two genes, ARID1A and ARID1B, survive only a quarter as long as patients without the changes. The discovery could eventually lead to early identification of patients with aggressive neuroblastomas who may need additional treatments.
Neuroblastomas affect nerve tissue throughout the body and are the most common, non-blood cancer in children. "These cancers have a wide spectrum of clinical outcomes, with some that are highly curable and others very lethal," says Victor Velculescu, M.D., Ph.D., professor of oncology and co-director of the Cancer Biology Program at Johns Hopkins. "Part of the reason for this variety in prognosis may be due to changes in the ARID1A and ARID1B genes."
Velculescu said these powerful "bully" genes were not identified in other gene sequencing studies of neuroblastoma, most likely because the Johns Hopkins-CHOP researchers used sequencing and analytical methods that looked for larger, structural rearrangements of DNA in addition to changes in the sequence of individual chemical base-pairs that form DNA. A report of their work appears in the Dec. 2 issue of Nature Genetics.
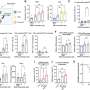
Of the 74 tumors in the study, 71 were analyzed for both rearrangements and base-pair changes. Cancer-specific mutations were found in a variety of genes previously linked to neuroblastoma, including the ALK and MYCN genes. In eight of the 71 patients, the investigators found alterations in the ARID1A and ARID1B genes, which normally control the way DNA folds to allow or block protein production.
The children with ARID1A or ARID1B gene changes had far worse survival, on average, than those without the genetic alterations—386 days compared with 1,689 days. All but one of these patients died of progressive disease, including one child whose neuroblastoma was thought to be highly curable.
The scientists were also able to detect and monitor neuroblastoma-specific genetic changes in the blood of four patients included in the study, and correlated these findings to disease progression.
"Finding cancer-specific alterations in the blood could help clinicians monitor patients for relapse and determine whether residual cancer cells remain in the body after surgery," says Mark Sausen, a Johns Hopkins graduate student and one of the lead scientists involved in the research.
The Johns Hopkins-CHOP team plans to conduct further studies in larger groups of patients to confirm the ARID1A-ARID1B correlation to prognosis.
More information: "Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma," Nature Genetics, advance online publication, Dec. 2, 2012. doi: 10.1038/ng.2493