Immunology research sheds new light on cell function, response
A Kansas State University-led study has uncovered new information that helps scientists better understand the complex workings of cells in the innate immune system. The findings may also lead to new avenues in disease control and prevention.
Philip Hardwidge, associate professor of diagnostic medicine and pathobiology, was the study's principal investigator. He and colleagues looked at the relationship between a bacterial protein and the innate immune system—a system of defensive cells that responds rapidly to an infection in a nonspecific manner.
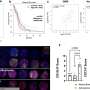
Among their findings, the researchers characterized a new protein that affects how cells in the innate immune system function and protect humans against invading bacteria such as E. coli O157:H7. The study, "NleB, a Bacterial Effector with Glycosyltransferase Activity, Targets GAPDH Function to Inhibit NF-kappaB Activation," was published in the most recent issue of the scientific journal Cell Host and Microbe. The National Institutes of Health's National Institute of Allergy and Infectious Diseases funded the study.
Hardwidge conducted the study with lead author Xiaofei Gao, a doctoral student at the University of Kansas Medical Center and now employed as a postdoctoral fellow at the Whitehead Institute; and with Thanh Pham and Leigh Ann Feuerbacher, postdoctoral research fellows in diagnostic medicine and pathobiology at Kansas State University. Colleagues at the University of Kansas Medical Center; the Institute of Infectiology in Muenster, Germany; and the Stowers Institute for Medical Research also contributed to the study.
The research team studied a bacterium that infects mice, named Citrobacter rodentium. The bacterium is similar to E. coli O157:H7, which causes diarrheal illness in humans. Both bacteria use the protein NleB to inhibit the innate immune system from fighting the bacteria.
"NleB is very important to the ability to cause disease," Hardwidge said. "Epidemiological and functional studies on E. coli and C. rodentium have shown that the presence of the NleB protein is associated with the ability of E. coli and C. rodentium to cause severe disease in humans and mice, respectively. But how the NleB protein did this was unknown."
According to Hardwidge, once bacteria such as C. rodentium and E. coli enter the body, the pathogens use a needle-like secretion apparatus to inject bacterial proteins into intestinal cells. Some of these proteins prevent the innate immune system from fighting the bacterium. One of these injected proteins is NleB.
Hardwidge and colleagues observed that the NleB protein binds with a protein in human cells named GAPDH. NleB modifies the GAPDH protein with a specific sugar molecule and prevents it from participating in a complex biochemical pathway that ultimately allows the innate immune system to respond efficiently to pathogens.
"The function of GAPDH in this pathway was less clear before we did these experiments," Hardwidge said. "GAPDH has well-known functions in the metabolism, but we observed that it also participates in how a cell responds to an infecting bacterium. We're very interested in the fact that this metabolic enzyme has apparently evolved also to be an important part of the innate immune system."
Hardwidge said that E. coli and C. rodentium using the NleB protein to target GAPDH and inhibit innate immunity is also an interesting finding, which will be characterized in greater detail in continuing studies.
With a more advanced understanding about how the innate immune system responds biochemically to invading bacteria—and how those bacteria suppress the response—scientists may be able to advance research and therapeutic drug development in other diseases, Hardwidge said. For example, cancers, Crohn's disease and Rheumatoid arthritis all are tied to overactive inflammation. In some cases, the same pathway in which GAPDH participates regulates the inflammation.
"The cell is so complicated, it's amazing that it even works at all, especially when you consider that it is three-dimensional and compartmentalized," Hardwidge said. "We have a general understanding about this important pathway that triggers a defensive response. But when you get into the details of how this pathway is regulated, we're still learning and understanding what exactly is going on. Now, low and behold, there is a new protein involved."