New study identifies unique mechanisms of antibiotic resistance
As public health authorities across the globe grapple with the growing problem of antibiotic resistance, Tufts University School of Medicine microbiologists and colleagues have identified the unique resistance mechanisms of a clinical isolate of E. coli resistant to carbapenems. Carbapenems are a class of antibiotics used as a last resort for the treatment of disease-causing bacteria, including E. coli and Klebsiella pneumonia, which can cause serious illness and even death. Infections involving resistant strains fail to respond to antibiotic treatments, which can lead to prolonged illness and greater risk of death, as well as significant public health challenges due to increased transmission of infection. The study, published in the April issue of Antimicrobial Agents and Chemotherapy, demonstrates the lengths to which bacteria will go to become resistant to antibiotics.
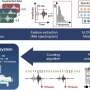
Resistance to carbapenems usually emerges through the acquisition of an enzyme, carbapenemase, which destroys the antibiotic intended to treat infection. Resistance may also block entry of the drug into the E-coli bacteria. The current research, led by corresponding author Stuart Levy, M.D., Professor of Molecular Biology & Microbiology and of Medicine and Director of the Center for Adaptation Genetics & Drug Resistance at Tufts University School of Medicine, sought to determine what made this particular clinical isolate of E. coli resistant to carbapenem in the absence of carbapenemase.
"The Centers for Disease Control and Prevention has documented a significant increase in Carbapenem-resistant Enterobacteriaceae (CRE) – so-called 'super bugs' that have been found to fight off even the most potent treatments," Levy said. "We knew that bacteria could resist carbapenems, but we had never before seen E. coli adapt so extensively to defeat an antibiotic. Our research shows just how far bacteria will go with mutations in order to survive."
Levy and his colleagues determined that the E. coli genetically mutated four separate times in order to resist carbapenems. Specifically, the isolate removed two membrane proteins in order to prevent antibiotics from getting into the cell. The bacteria also carried a mutation of the regulatory protein marR, which controls how bacteria react in the presence of antibiotics. The isolate further achieved resistance by increasing expression of a multidrug efflux pump. Moreover, the researchers discovered that the E. coli was expressing a new protein, called yedS, which helped the drug enter the cell, but whose expression was curtailed by the marR mutation. yedS is a normally inactive protein acquired by some E. coli that affects how the drug enters the bacterial cell. It is generally expressed in bacteria through a mutation.
According to the Centers for Disease Control and Prevention, CRE germs have increased from 1% to 4% in the United States over the last decade. Forty-two states report having identified at least one patient with one type of CRE. Approximately 18% of long-term acute care hospitals in the United States and 4% of short-stay hospitals reported at least one CRE infection in the first half of 2012.
The clinical isolate of E. coli studied by Levy and his colleagues came from the sputum of a patient at Peking Union Medical College Hospital in Beijing, China, where three of the study authors are on the faculty. Drug resistance is a particularly serious public health concern in China, antibiotics are overprescribed and used widely in the livestock and farming industries.
"The first quinolone-resistant strains of bacteria came out of China, where we see that the drugs of last resort begin being used, because the other drugs don't work after so much overuse," Levy said.
More information: Warner, D.M., Yang, Q., Duval, V., Chen, M., Xu, Y., Levy, S.B. (2013). Involvement of MarR and YedS in Carbapenem Resistance in a Clinical Isolate of Escherichia coli from China. Antimicrob. Agents Chemother., 57(4), 1935-1937. doi: 10.1128/AAC.02445-12










.jpg)