'RNA sponge' mechanism may cause ALS/FTD neurodegeneration
The most common genetic cause of both ALS (amyotrophic lateral sclerosis) and FTD (frontotemporal dementia) was recently identified as an alteration in the gene C9orf72. But how the mutation causes neurodegenerative disease appeared mysterious. Researchers at Emory University School of Medicine have demonstrated that this ALS/FTD mutation may be harmful because it creates an "RNA sponge," soaking up an important regulatory protein that binds RNA.
The results were published online Monday in the Proceedings of the National Academy of Sciences, Early Edition.
"We think that the RNA itself is part of the disease mechanism," says co-author Thomas Wingo, MD, assistant professor of neurology at Emory University School of Medicine. "In both cell culture and fruit flies, we've been able to show that we can add back the protein depleted by the RNA and ameliorate the problem."
The finding provides insight into the mechanism of disease in ALS and FTD, both in cases where C9orf72 is altered and in other cases. It suggests some forms of ALS/FTD may have common elements with other neurodegenerative disorders caused by noncoding repeats, such as myotonic dystrophy, spinocerebellar ataxia and fragile X-associated tremor/ataxia syndrome.
The senior author is Peng Jin, PhD, professor of human genetics at Emory University School of Medicine. The first authors are a former graduate student, Zihui Xu, PhD, now at Huazhong University of Science and Technology in China, and research specialist Mickael Poidevin.
ALS is a fatal disease in which motor neurons in the brain and spinal cord degenerate. As the illness progresses, patients lose the ability to walk, talk and breathe. FTD is a form of dementia in which the patients primarily experience deterioration in behavior, personality or language.
Many neurologists and researchers consider these conditions as sharing clinical and pathological features. Mutations in several genes have been linked to both ALS and FTD, which suggests that they have a common mechanism. However, most cases are considered sporadic, meaning that they don't have a clear family history.
In 2011, a mutation was identified within C9orf72 as the most common genetic cause of ALS and FTD, accounting for 5 to 7 percent of cases of each disease. The mutation doesn't seem to affect the protein encoded by C9orf72. Instead, it expands a block of repetitive DNA, so that the sequence "GGGGCC" is repeated hundreds of times outside the parts of the gene that encode protein.
This looks similar to "noncoding repeat" expansions responsible for other neurodegenerative disorders such as myotonic dystrophy, spinocerebellar ataxia and fragile X-associated tremor/ataxia syndrome.
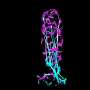
Some researchers have reported that the GGGGCC repeat produces RNA that is translated into an unusual protein that aggregates in cells. These protein aggregates are thought to be toxic to neurons. Emory researchers have an alternative explanation: the GGGGCC repeat RNA itself is toxic.
The Emory team tested the effects of producing the GGGGCC repeat RNA, which proved toxic to cultured mammalian neuronal cells and caused neurodegeneration in fruit flies. When the repeat RNA was produced in flies' motor neurons, the flies had reduced motor activity.
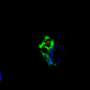
The repeat RNA appears to be harmful because, when overproduced, it sequesters a protein called Pur alpha, which sticks to the GGGGCC repeats. Scientists had previously found that Pur alpha is necessary for neuronal development and is involved in the transport of RNA within neurons. Making neuronal cells or flies produce more Pur alpha to compensate reverses the toxicity caused by the RNA, the Emory scientists found.
"For ALS and FTD, this suggests a disease-fighting strategy of targeting either the toxic RNA itself or its interaction with Pur alpha," Jin says. "It also hints that there is a common RNA-based mechanism contributing to several neurodegenerative diseases."
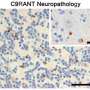
Sequestering Pur alpha appears to cause its redistribution within the neurons of patients affected by ALS/FTD. The Emory team was able to detect clumpy "inclusions" containing Pur alpha in samples of brain tissue from individuals with C9orf72 mutations – and also in other individuals with FTD but without C9orf72 mutations.
More information: Z. Xu, M. Poidevin, X. Li, Y. Li, L. Shu, D.L. Nelson, H. Li, M. Gearing, T.S. Wingo and P. Jin. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes, PNAS, 2013.