New method for predicting cancer virulence developed
A new way of tackling cancer and predicting tumor virulence are has been reported by a French team of scientists from the Institut Albert Bonniot de Grenoble including researchers from CNRS, Inserm and Université Joseph Fourier. The scientists have shown that, in all cancers, an aberrant activation of numerous genes specific to other tissues occurs. For example, in lung cancers, the tumorous cells express genes specific to the production of spermatozoids, which should be silent. This work, published on 22 May 2013 in Science Translational Medicine, suggests that identifying the genes that are abnormally activated in a cancer makes it possible to determine its virulence with great accuracy. This study represents an original concept that will allow cancer patients to be given an accurate diagnosis as well as personalized care.
All the cells in our body have the same genes. However, their specialization leads them to activate some and repress others. In a cancerous cell, the mechanisms that allow a cell to activate or silence genes are damaged. The scientists have shown that, in all cancers, a sort of "identity crisis" is observed in cancerous cells: in the organs or tissues in which a tumor develops, genes specific to other tissues or to other stages of the development of the organism express themselves in an aberrant manner. Until now, this aspect had only been partially studied.
By focusing on these genes that are "awakened" in tumors, the researchers have shown that in almost all cancers, tens of specific genes in the germline and the placenta are aberrantly activated. This represents a very interesting source of potential biomarkers for characterizing tumors.
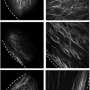
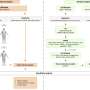
In order to explore the implications of these aberrant activations and their significance, the scientists concentrated their efforts on lung cancer. They examined the tumors of nearly 300 patients at the CHU de Grenoble. Over a ten year period, the physicians documented patients' records and conserved and annotated tumors after surgical resection. The expression of all human genes was analyzed in these tumors and correlated with different clinical parameters.
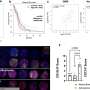
Among the genes aberrantly expressed in lung cancers, they discovered 26 whose activation is associated with particularly aggressive cancers: when these genes are expressed, the cancer is extremely virulent. The researchers are thus able to predict, at the diagnosis stage, which cancers have a high risk of recurrence and a fatal prognosis, even in cases where the tumor is adequately treated, at an early stage of its development. These high-risk cancers exhibit increased proliferative abilities and a facility to "hide" from the body's immune system.
This work provides a proof of principle for a new approach in the study and treatment of cancer: the aberrant expression in a tissue or organ of genes specific to other tissues could become a new tool for establishing a prognosis and personalizing therapeutic care. From a more fundamental viewpoint, the researchers still need to explain the relationship between the aberrant expression of these genes and the virulence of the cancer. An approach similar to that tested in lung cancer could be extended to virtually all types of cancer, opening very broad perspectives regarding the exploitation of these findings.
More information: Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers, Sophie Rousseaux et al. Science Translational Medicine, 22 May 2013