Novel RNA-based classification system for colorectal cancer
A novel transcriptome-based classification of colon cancer that improves the current disease stratification based on clinicopathological variables and common DNA markers is presented in a study published in PLOS Medicine this week. (A transcriptome is all RNA produced by a population of cells.) Pr. Pierre Laurent-Puig and colleagues from INSERM in Paris, France used genetic information from a French multicenter study supported by the "Ligue contre le cancer" to identify a standard, reproducible molecular classification based on gene expression analysis of colorectal cancer. The authors also assessed whether there were any associations between the identified molecular subtypes and clinical and pathological factors, common DNA alterations, and prognosis.
Cancer of the large bowel (colorectal cancer) is the third most common cancer in men and the second most common cancer in women worldwide. Despite recent advances in the screening, diagnosis, and treatment of colorectal cancer, an estimated 608,000 people die every year from this form of cancer—8% of all cancer deaths. The prognosis and treatment options for colorectal cancer depend on five pathological stages (0–IV), each of which has a different treatment option and five year survival rate, so it is important that the stage is correctly identified. Unfortunately, pathological staging fails to accurately predict recurrence (relapse) in patients undergoing surgery for localized colorectal cancer.
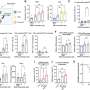
The authors used genetic information from a cohort of 750 patients with stage I to IV colorectal cancer who underwent surgery between 1987 and 2007 in seven centers in France. The researchers identified relevant clinical and pathological staging information for each patient from the medical records and calculated recurrence-free survival (the time from surgery to the first recurrence) for patients with stage II or III disease. Using these methods, the authors classified colon cancer samples into six molecular subtypes (based on gene expression data). Importantly, the researchers found that the six identified subtypes were associated with distinct clinical and pathological characteristics, molecular alterations, specific gene expression signatures, and deregulated signaling pathways. In the prognostic analysis, the researchers found that patients whose tumors were classified in clusters C1-C3 or C5 had a relative 50% greater likelihood of relapse-free survival than those with clusters C4 or C6, even after adjusting for age, sex, cancer stage, and Oncotype recurrence score (hazard ratio, 1.5, 95% confidence interval 1.1-2.1, P=0.0097).
Strengths of the study include the large well-characterized colon cancer samples included from multiple centers and validation using an independent dataset. However, this study was retrospective and did not include some known predictors of colorectal cancer prognosis, such as tumor grade and number of nodes examined. The significance and robustness of the prognostic classification requires further confirmation with large prospective patient cohorts.
The authors conclude, "We describe the first, to our knowledge, robust transcriptome-based classification that improves the current disease stratification based on clinicopathological variables and common DNA markers. The biological relevance of these subtypes is illustrated by significant differences in prognosis. This analysis provides possibilities for improving prognostic models and therapeutic strategies."
More information: Marisa L, de Reynie`s A, Duval A, Selves J, Gaub MP, et al. (2013) Gene Expression Classification of Colon Cancer into Molecular Subtypes: Characterization, Validation, and Prognostic Value. PLoS Med 10(5): e1001453. doi:10.1371/journal.pmed.1001453