Homing in on developmental epigenetics
Germ cells have unique molecular features that enable them to perform the important task of transmitting genetic information to the next generation. During development from their embryonic primordial state, germ cells are thought to be reprogrammed by epigenetic modification of their DNA. However, due to the significant technical challenges associated with epigenetic analysis of rare cells such as primordial germ cells, the exact nature and effect of such epigenetic changes remains poorly understood. Kuniya Abe, Rieko Ikeda and colleagues from the RIKEN BioResource Center have now developed a method that enables as few as 100 cells to be analyzed for epigenetic changes, providing new insight into these processes.
Epigenetic changes regulate gene expression without altering the underlying DNA sequence. They involve highly specific chemical modifications of DNA and DNA-bound proteins that activate or deactivate individual or related sets of genes. DNA methylation is one such epigenetic mechanism, and the technique developed by Abe's team is an adaptation of an existing microarray-based assay method for detecting DNA methylation. "Our modification of the state-of-the-art approach might be useful for basic research as well as diagnostic applications when less than a nanogram of genomic DNA is available," says Abe.
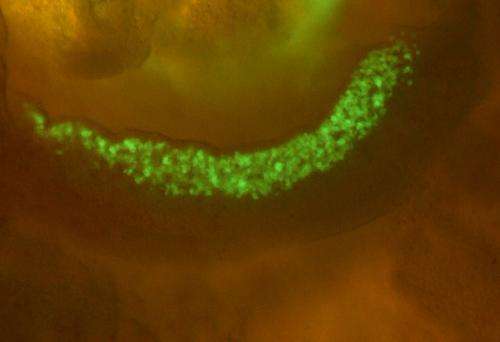
Using their technique on embryonic mouse cells, the researchers compared the extent to which the genomic DNA of somatic cells, stem cells, and both male and female germ cells at different stages of development is tagged with methyl groups. Looking specifically at germ cells (Fig. 1), they found that large stretches of DNA on the X chromosome of male germ cells were depleted of methyl groups. These large 'hypomethylated' domains frequently included genes with germ cell- or testis-specific expression, including cancer testis antigen (CTA) genes, which are expressed in male germ cells and cancer cells, but not in normal somatic cells. Genes in these regions were also expressed despite the presence of a histone modification that normally suppresses gene expression.
"The mechanistic basis for the relationship between hypomethylation and the expression of CTA genes remains to be established," says Abe. "We suspect that a change in nuclear architecture that controls the accessibility of the genes to the transcriptional apparatus might be involved."
Abe also notes that if CTA gene expression does indeed contribute to oncogenesis, then understanding the epigenetic regulation of these genes in germ cells could provide insight into the molecular events that cause certain types of cancer.
More information: Ikeda, R., et al. Large, male germ cell-specific hypomethylated DNA domains with unique genomic and epigenomic features on the mouse X chromosome, DNA Research, 15 July 2013. dx.doi.org/10.1093/dnares/dst030