Fate of new genes cannot be predicted
New versions of genes, called alleles, can appear by mutation in populations. Even when these new alleles turn the individuals carrying them more fit to survive and reproduce, the most likely outcome is that they will get lost from the populations. The theory that explains these probabilities has been postulated by the scientist J.B.S. Haldane almost 90 years ago. This theory has become the cornerstone of modern population genetics, with studies on adaptation to novel environments and conservation of species, for example, being based on it. However, until now there were no explicit experimental tests of this theory.
The research team led by Henrique Teotónio, at the Instituto Gulbenkian de Ciência (IGC, Portugal), in collaboration with Isabel Gordo, also from the IGC, has now experimentally tested Haldane's theory. By performing competition tests in roundworms, they have confirmed this theory for the introduction of a new beneficial allele in a population. However, the researchers found that this theory cannot predict the ultimate fate of the allele. This study, published in the latest issue of the scientific journal Nature Communications, contributes to a better comprehension of how a population can evolve, with implications for studies on how species adapt to changing environments or species conservation.
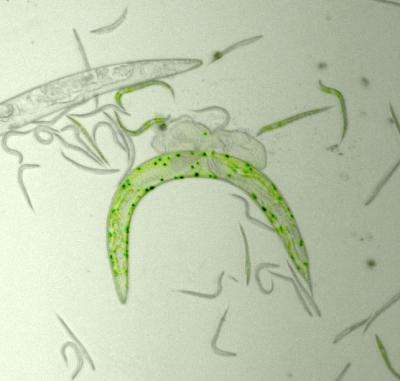
Haldane's theory could be experimentally tested because the researchers set up the ideal conditions with the roundworm Caenorhabditis elegans (C. elegans). These tiny organisms primarily reproduce by self-fertilization, which assures that genetic identity is maintained. Also, these organisms reproduce in a short timeframe, making possible the study of several generations. Taking advantage of C. elegans' characteristics, Ivo Chelo and Judit Nédli used two lines of C. elegans to establish competition assays and see which individuals could survive and reproduce better.
Chelo and colleagues started by verifying Haldane's theory at the stage of introducing a new allele in the population. They introduced 2 or 5 individuals from one line in a population of 1000 individuals from another line, and tested how those individuals invaded the population. They found out that when the invaders were more fit the introduction in the population of more individuals would lower their probability of extinction. They also confirmed that the probability of extinction is higher for deleterious alleles than for beneficial alleles. Nonetheless, less fit individuals could be kept in the population for a few generations at high frequencies, the more so if population sizes were small. The validity of Haldane's theory had been proved for the initial phases of invasion.
Then, the research team addressed the probability of fixation of a beneficial allele in the population, i.e., to have all individuals of the population carrying the new allele. They repeated the competition experiments between the two same lines of C. elegans, but this time used an initial higher number of invading individuals, to mimic a population in which the beneficial allele was already established. The researchers observed that the adaptive value of each allele, i.e., whether it behaves as beneficial or deleterious, depended on its frequency in the population. If its frequency was higher than 5% (when more than five different individuals in a population of 100 individuals), the allele was perceived as deleterious and it started to be eliminated by natural selection. But when the frequency was less than 5%, the allele was beneficial. The result of these complex dynamics is that genetic diversity could be maintained indefinitely, without one allele or the other ever being fixed in the population.
Ivo Chelo explains: "Our data suggests that the value a new allele brings to the individuals is not fixed. Populations are dynamic and complex with plenty of interactions between individuals and between these and the environment. Initial stages when the new alleles appear cannot tell us what the effects of the alleles will be a few generations later, when the population has already changed."
Henrique Teotónio adds: "To our knowledge, this is the first time anyone was able to directly test Haldane's theory. We have proved it correct for the initial stages, when a new allele appears in a population. But our results show that further empirical work and more theoretical models are required to accurately predict the fate of that allele over long time spans".
More information: Nature Communications 4:2417 DOI: 10.1038/ncommsS3417