Mapping mechanisms to medications
By charting shifts in gene expression, researchers can gain deeper insights into drug mechanisms and side effects.
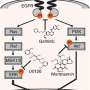
The timing of gene activity is primarily controlled by regulatory sequences known as promoters, which contain various binding sites for different proteins that help switch genes on or off. The majority of these binding sites are likely to appear in numerous different promoters, such that a single cellular signal can orchestrate the activity of multiple genes in parallel.
Researchers led by Hideya Kawaji from the RIKEN Preventive Medicine and Diagnosis Innovation Program and Harukazu Suzuki from the RIKEN Center for Life Science Technologies have now devised a strategy for quantifying how different drugs and drug candidates modulate activities across the entire genome within cells. In principle, this information could be utilized to predict the efficacy of drug candidates toward a given disease or the risk of unwanted side effects.
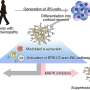
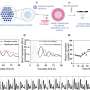
Their approach is based on cap analysis of gene expression (CAGE), a method originally developed by RIKEN scientists to measure changes in gene output. By mapping these shifts back to their context in the genome, the researchers were able to identify exactly which promoters were being modulated in response to different compounds. In an initial study, they treated cultured breast cancer cells with wortmannin, gefitinib and U0126—three compounds with known antitumor effects. This CAGE-based approach, in combination with a new generation of DNA sequencing technology, proved both reproducible across multiple experiments and highly sensitive, detecting even the subtle perturbations in promoter function that arose from low doses of a given drug.
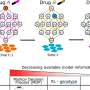
For each of these drugs, Kawaji, Suzuki and their colleagues were able to identify specific effects on more than 100 different promoters. The results were consistent with the mode of action and targeted genes previously reported in extensive studies of these compounds. For example, the signaling pathway targeted by gefitinib in turn modulates two separate pathways inhibited by wortmannin and U0126, and modeling based on the CAGE data confirmed that the great majority of promoters affected by these latter two drugs could indeed account for most of the effects observed for gefitinib.
"This is a very important finding," explains Suzuki, "because it means that we can now predict the targets and mechanisms of new drugs using the profiles of known drugs."
The researchers' approach could also provide a deeper understanding of minor or 'off-target' drug effects that are unrelated to the disease being treated but nevertheless exert potentially meaningful physiological effects. "This is very important in many aspects of drug research, such as side effects and synergic effects with other drugs," says Suzuki.
More information: Kajiyama, K., Okada-Hatakeyama, M., Hayashizaki, Y., Kawaji, H. & Suzuki, H. Capturing drug responses by quantitative promoter activity profiling. CPT: Pharmacometrics & Systems Pharmacology 2, e77 (2013). dx.doi.org/10.1038/psp.2013.53