Helping cancer researchers make sense of the deluge of genetic data
A newly improved internet research tool is helping cancer researchers and physicians make sense out of a deluge of genetic data from nearly 100,000 patients and more than 50,000 mice.
The tool, called the Gene Expression Barcode 3.0, is proving to be a vital resource in the new era of personalized medicine, in which cancer treatments are tailored to the genetic makeup of an individual patient's tumor.
Significant new improvements in the Gene Expression Barcode 3.0 are reported in the January issue of the journal Nucleic Acids Research, published online ahead of print.
Senior author is Michael J. Zilliox of Loyola University Chicago Stritch School of Medicine. Zilliox is co-inventor of the Gene Expression Barcode.
"The tool has two main advantages," Zilliox said. "It's fast and it's free."
The Gene Expression Barcode is available at a website http://barcode.luhs.org/ designed and hosted by Loyola University Chicago Stritch School of Medicine. The website is receiving 1,600 unique visitors per month.
Knowing how a patient's cancer genes are expressed can help a physician devise an individualized treatment. In a tumor cell, for example, certain genes are turned on (expressed) while other genes are turned off (unexpressed). Also, different types of cancer cells have different patterns of gene expression. Genes are expressed through RNA, a nucleic acid that acts as a messenger to carry out instructions from DNA for making proteins.
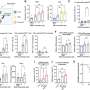
Research institutions have made public genetic data from nearly 100,000 patients, most of whom had cancer, and more than 50,000 laboratory mice. In raw form, however, these data are too unwieldy to be of much practical use for most researchers. The Gene Expression Barcode applies advanced statistical techniques to make this mass of data much more user-friendly to researchers.
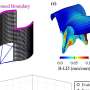
The barcode algorithm is designed to estimate which genes are expressed and which are unexpressed. Like a supermarket barcode, the Gene Expression Barcode is binary, meaning it consists of ones and zeros—the expressed genes are ones and the unexpressed genes are zeroes.
Zilliox co-invented the Gene Expression Barcode, along with Rafael Irizarry, PhD. (At the time, Zilliox and Irizarry were at Johns Hopkins University.) Zilliox joined Loyola in 2012, and Irizarry now is at the Dana Farber Cancer Institute. Zilliox and Irizarry first reported the Gene Expression Barcode in 2007. In 2011, they reported an improved 2.0 version. The Barcode already has been cited in more than 120 scientific papers, and the new 3.0 version will make it even easier and faster for researchers to use, Zilliox said.
The Gene Expression Barcode is supported by funding from the National Institutes of Health and Loyola institutional funds.
In addition to Zilliox and Irizarry, co-authors of the article describing the Barcode 3.0 version are Matthew McCall of the University of Rochester, Harris Jaffee of Johns Hopkins University, Susan Zelisko of Loyola University Chicago Stritch School of Medicine and Neeraj Sinha and Guido Hooiveld of Wageningen University.
In the paper, the authors thank Joseph Koral, Baimei Guo, Corey Sartin and Ron Price of Loyola's Informatics and Systems Development for their computational support.
The article is titled "The Gene Expression Barcode 3.0: Improved Data Processing and Mining Tools"