Algorithm reconstructs lineages of competing cancer cells within individual tumours
Researchers at the Wellcome Trust Sanger Institute have developed a method to characterise the main lineages of cells that are present in a tumour and their prevalence over time following chemotherapy. Aiming to reconstruct the genetic profiles of cellular lineages within the tumour, the team created a new computational algorithm named cloneHD which makes use of whole-genome DNA sequencing from mixed populations.
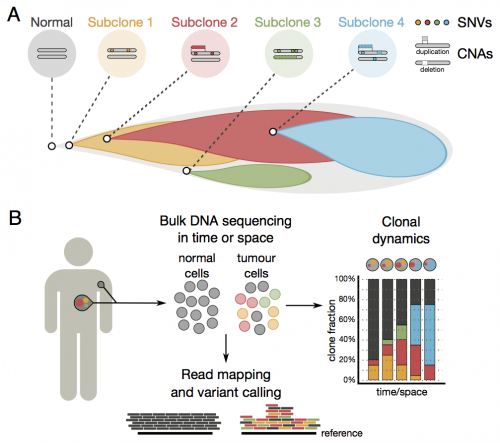
Cells in a tumour originate from a normal cell giving rise to billions of malignant, genetically diverse cells. Genome-wide approaches to study clonal heterogeneity are hampered by limited resolution of subpopulations, since short-read sequencing of mixed cell populations does not yield a direct readout of their clonal identity. There is mounting evidence for genetic heterogeneity in the majority of cancer types, which can translate in a differential response to treatment.
To address this problem, the algorithm uses short-read sequence data that can be related to different types of aberrations within all the genomes in a population to carry out its analysis. By combining these data the researchers can identify and describe the major lineages of genetically distinct cancer cells, also known as clones. The algorithm classifies cells into groups, discriminating different cell populations that belong to each of the clonal lineages. At the genetic level, the algorithm can assign aberrations by clone: somatic changes to single letters and copy-number changes in the genetic code, as well as allelic imbalances in the germline caused by these events.
"Our cloneHD algorithm helps us generate useful insights into cancer development and the effects of treatment, by delineating cell lineages within the tumour and their progression over time." says Dr Ville Mustonen, senior author from the Sanger Institute.
To test that the algorithm can trace clonal dynamics in cancer, the researchers retrospectively explored temporal changes in a case study of a patient with chronic lymphocytic leukaemia, from whom sequence data had been collected at five different time points during the course of treatment. Such retrospective analysis is helpful to better understand tumour evolution. They further performed a mimic of real-time monitoring by progressively adding more samples to see if prospective analyses are of potential clinical relevance.
"The algorithm allowed us to see that the dominant lineage of tumour cells diminished in response to treatment, but that another lineage of drug-resistant cells had started to rise in numbers," says Dr Andrej Fischer, first author from the Sanger Institute. "It is possible that, using this approach in conjunction with near real-time data collection, clinicians will be able to identify when a treatment is starting to fail."
In the near future, the team will test the algorithm and its potential clinical applications using larger data sets and a wider range of cancer types. Beyond cancer, a quantitative understanding of clonal dynamics translates to other organisms facing environmental stresses, such as bacteria, parasites or viruses.
More information: "High-definition reconstruction of clonal composition in cancer." Fischer A, Vázquez-García I, Illingworth CJR, Mustonen V. Cell Reports 2014. DOI: 10.1016/j.celrep.2014.04.055