Study identifies potential genes associated with the most common form of liver damage
In a first-of-its-kind exploratory study, the Translational Genomics Research Institute (TGen) has identified a potential gene associated with the initiation of the most common cause of liver damage.
Nonalcoholic fatty liver disease (NAFLD) is the most common cause of liver damage. In this study, published in the September edition of Translational Research, TGen scientists sequenced microRNAs (miRNAs) from liver biopsies, spelling out their biochemical molecules to identify several potential gene targets associated with NAFLD-related liver damage.
The miRNAs—RNA molecules that regulate gene expression—were obtained from liver biopsies of 30 female candidates for gastric bypass surgery: 15 with, and 15 without, NAFLD liver damage.
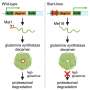
Using the most advanced technology to refine the data, researchers identified several potential gene targets associated with NAFLD-related liver damage. Specifically, they found that a particular miRNA called miR-182 produced a strong association with a protein called FOX03.
"Because of the known role of miR-182 in mechanisms related to liver cancer, we sought to investigate this miRNA in NAFLD-related liver damage by looking at relevant target genes," said TGen Research Associate Fatjon Leti, the study's lead author. "We found that levels of FOX03, which has been implicated in liver metabolism, to be significantly decreased."
Importantly, researchers observed a significant suppression of FOX03 protein levels in damaged livers, compared to those without liver damage, suggesting a potential role for this gene in the initiation of liver disease.
"These finding support a role for liver miRNAs in the disease development of NAFLD-related damage, and yield possible new insight into the molecular mechanisms underlying the initiation and progression of liver damage and eventual liver failure," said Dr. Johanna DiStefano, Professor and Director of TGen's Diabetes, Cardiovascular & Metabolic Diseases Division, and the study's senior author.
"To our knowledge, this is the first study to apply a high-throughput sequencing approach to the investigation of liver miRNAs in NAFLD-related liver damage," DiStefano said.
The authors cite limitations in this particular study: The study's small sample, and the fact that all 30 patients were obese females who were candidates for gastric bypass surgery, which may have limited the data. They were selected because obesity is a significant risk factor for NAFLD outcomes, and because of the value of obtaining unbiased liver samples.
"We consider this study an exploratory one, and we acknowledge that validation in a larger, independent dataset will be necessary to confirm our findings," DiStefano said. "The results reported here do not allow us to make specific conclusions about miRNAs and biological pathways. Additional studies will be necessary to confirm the role of specific miRNAs in liver damage."
More information: High-throughput sequencing reveals altered expression of hepatic microRNAs in nonalcoholic fatty liver disease-related fibrosis, Translational Research, 2015.