New pathway to differentiation found, shedding light on some cancers
In humans the differentiation of stem cells into hundreds of specialized cell types is vital. Differentiation drives development from fertilized egg to a newborn, and it underlies the continuous replacement of the 5 billion cells that die every hour in an adult. On the downside, mutations in differentiation pathways of different cell types can be drivers of cancers.
Xinyang Zhao, Ph.D., and colleagues Li Zhang, Ngoc-Tung Tran and Hairui Su at the University of Alabama at Birmingham and at seven other institutions have discovered a new mechanism of differentiation, as studied in megakaryocytes, the blood cells responsible for platelet production.
This mechanism may offer new research and therapeutic approaches to blood malignancies as well as solid tumors such as colon and breast cancers. Since two of the key proteins in the pathway—PRMT1 and RBM15—are expressed in all tissues, they may also regulate differentiation in other tissues, especially those where RBM15 has been shown to be essential for development, including the heart, spleen and placenta.
The mechanism that controls the decision of a progenitor cell to take one fork or another on the road to cell specialization is key to understanding differentiation. Since PRMT1 and RBM15 are highly conserved evolutionarily among plants, amphibians, fish, birds and mammals, the new pathway may be involved in those diverse organisms. Each organism has numerous specialized cell types, but all those specialized cells arise from less specialized progenitor cells that can branch into a variety of specialized cells. An example of this in humans is the blood stem cell in the bone marrow, which can specialize into a dozen different blood cells such as red blood cells, neutrophils or natural killer cells. Billions of specialized blood cells are created every day.
The work by Zhao and his colleagues, published online in advance of print in eLife, has taken four years.
The pathway is complex to describe, but its ultimate effect is an alternative splicing of messenger RNAs—particularly the messenger RNAs of master regulators called transcription factors. These factors control the reading of genetic information from DNA genes and act as switches in all living organisms to control gene expression. Though a nerve cell and a muscle cell have the same genomes, for example, different gene expression gives each its distinct qualities.
Pathway details
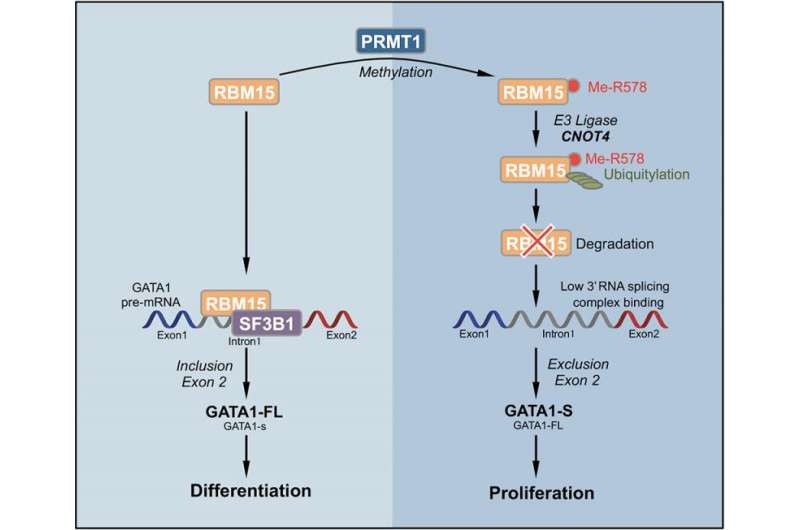
The pathway starts with the methyltransferase enzyme PRMT1. This enzyme attaches a methyl group onto specific arginine amino acid residues of target proteins. The Zhao team screened for proteins that were tagged with methyl groups by PRMT1 and selected one of them—the RNA-binding protein RBM15—for further study. RBM15 was of interest because a mutant fusion of RBM15 and MKL1 proteins is associated with acute megakaryoblastic leukemia.
In experiments, Zhao and colleagues found that when PRMT1 levels are high in a cell, a greater proportion of RBM15 is tagged with methyl groups on certain arginine residues. This tagging causes a ligase called CNOT4 to mark RBM15 with another tag, ubiquitin—the well-known signal that marks a protein for transport to the cell's garbage removal machinery, where the protein is degraded and recycled. Zhao's team found that the methyl-tagged RBM15 proteins rapidly disappeared, even though the amount of RBM15 mRNA did not change. Thus, the expression levels of PRMT1 inversely affected the amount of RBM15 protein. While protein ubiquitylation was previously known to be triggered by phosphorylation and lysine methylation, this triggering by arginine methylation was unknown.
When the concentration of RBM15 protein is low, the megakaryocytic progenitor cells cannot move forward to differentiation. When the concentration of RBM15 is high enough, the progenitor cells undergo differentiation into mature megakaryocytes, the specialized blood cells that produce blood-clotting platelets in mammals.
To understand how this switch functions, a brief explanation of RNA splicing is needed. Inside the cell nucleus, a gene is encoded by a sequence of DNA. This gene sequence is copied into a strand of RNA, which is then processed through a ribosome to construct the protein encoded by the DNA. The ribosome's reading of the sequence of RNA bases tells the ribosome which sequence of amino acids, one by one, get added to build the protein. But there is a complication: The DNA gene sequence, and thus the RNA copy also, contain non-reading inserts called introns. Before the ribosome can make the protein, these introns must be cut out by RNA splicing enzymes. As they remove introns, the splicing enzymes join together the ends of the reading sequences, called exons. This creates the proper messenger RNA for the ribosome.
Thus a gene with three exons and two introns is analogous to this sentence: "The quick red xxxxxxxxxx fox jumped over yyyyyyyyyy the lazy brown dog." After splicing, the message would read, "The quick red fox jumped over the lazy brown dog." These exons need to be spliced together as the introns are removed.
Zhao and colleagues found that RBM15 RNA-binding protein binds to intron regions of the pre-messenger RNA for genes known to be important in megakaryocyte differentiation, including three transcription factors such as RUNX1, GATA1 and TAL1 known to be important for normal and abnormal hematopoiesis. RBM15 appears to recruit the splicing factor SF3B1 to correctly splice the exons. When RBM15 is low, one or more exons are not correctly spliced. Thus, this is a new mechanism for cell differentiation, initiated by methylation of RNA-binding proteins.
"The regulation of alternative splicing by RBM15 through SF3B1 is an exciting and novel pathway that clearly participates in the decision of a megakaryocyte to grow or differentiate," said John Crispino, Ph.D., professor of hematology/oncology, and biochemistry and molecular genetics, at the Northwestern University Feinberg School of Medicine. "These findings suggest that modulation of RBM15 activity by suppressing PRMT1 activity may change the splicing pattern of megakaryocytic tumor cells and facilitate their differentiation."
Broader role?
RBM15 may have broader functions in cells, say Zhao and colleagues. They found that RBM15 binds directly to the pre-messenger RNA of 1,257 genes. Among them are genes involved in metabolic regulation. In agreement with this finding, Zhao and colleagues found that overexpression of PRMT1 or reduced expression of RBM15 enhanced the creation of more mitochondria, the powerhouses of the cell. In collaboration with the Memorial Sloan Kettering Cancer Center lab of Minkui Luo, Ph.D., Zhao's group has further identified metabolic pathways regulated by PRMT1 in leukemia cells. These data, in a manuscript under preparation, will further link tumorigenesis to metabolic pathways.
Alternative RNA splicing is attracting attention in cancer research. The splicing factor SF3B1 contains mutations in more than 70 percent of myelodysplastic syndrome patients and 20 percent of chronic lymphocytic leukemia patients, and mutated SF3B1 also appears in other types of hematological malignancies. Understanding the PRMT1-RBM15 axis can shed new light on SF3B1-mutated hematological malignancies and may lead to targeting PRMT1 as a novel therapy for myelodysplastic syndromes. In a long-term collaboration with Y. George Zheng, Ph.D., at the University of Georgia, Zhao's group has been testing PRMT1 inhibitors.
More information: Li Zhang et al. Cross-talk between PRMT1-mediated methylation and ubiquitylation on RBM15 controls RNA splicing, eLife (2015). DOI: 10.7554/eLife.07938