A new biomarker for nerve cell damage
Scientists at the German Center for Neurodegenerative Diseases, the Hertie Institute for Clinical Brain Research and the University of Tübingen have identified proteins in the blood and cerebrospinal fluid that reflect nerve cell damage. The results of the study, published in the journal Neuron, suggest that the concentration of these 'neurofilament light chain proteins' could provide information about the progression of neurodegenerative diseases and the effects of treatment. Such a biomarker would be valuable for developing therapies.
"The results of our research indicate that disease progression can be tracked by monitoring the concentration of neurofilament light chain proteins. According to our study this is possible in both animal models and humans," says Mathias Jucker, who leads a research group at the DZNE's Tübingen site and is also a director of the Hertie Institute. "We have also found that the measured levels react sensitively when pathological hallmarks in the brain are influenced experimentally. It may thus be possible to evaluate the effect of a treatment by measuring the concentration of light neurofilaments, both in preclinical laboratory studies and in clinical trials. Such a biomarker would be a major benefit for developing treatments."
Parts of the cytoskeleton
"Neurofilament light chain proteins" are parts of the cytoskeleton that gives nerve cells shape and stability. These thread-like molecules are therefore mainly located inside cells, but may be released due to damage.
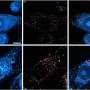
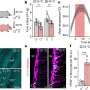
Professor Jucker and his colleagues - including Mehtap Bacioglu, first author of the current publication - took this known fact as the basis for investigating the concentration of neurofilament light chains proteins in the blood and cerebrospinal fluid (liquor). For this, they looked at mice that showed the typical features of neurodegenerative diseases, namely deposits of aggregated "alpha-synuclein", "tau" or "beta-amyloid" protein in their brains. Such deposits are associated with nerve cell damage. Besides, the scientists examined biomaterials from patients with Alzheimer's, Parkinson's and other neurodegenerative diseases.
Sensitive measurements
In mice, a close association was found between the concentration of neurofilament proteins in the liquor and blood. Moreover, the more advanced the brain damage, the higher the measured levels. If the neurological lesions were induced or inhibited protein levels increased or dropped accordingly. In patients, blood and liquor readings also correlated strongly. Furthermore, levels were higher than in healthy people.
A tool for developing treatments
"The special potential of this biomarker comes from the fact that it is significant in both animals and humans. Therefore, the results from animal models can be translated into clinical studies and their findings may be directly compared. This is critical for the development of new treatments," says Jucker. "What's more, we don't have to rely on withdrawals of liquor. The lumbar puncture required to obtain cerebrospinal fluid can be stressful for the person undergoing it. Our study shows that blood levels also provide information about neurodegeneration in the brain, because the concentrations of the neurofilaments in the blood and cerebrospinal fluid are closely coupled. A simple blood sample may therefore be sufficient when performing clinical studies on humans."
More information: Neurofilament light chain in blood and CSF as marker of disease progression in mouse models and in neurodegenerative diseases, Neuron, DOI: 10.1016/j.neuron.2016.05.018