Researchers identify genome-modifying enzyme linked to Rett Syndrome

People with Rett Syndrome, a rare and debilitating neurodevelopmental disorder, exhibit many autism-like traits. Now researchers at MIT have identified a protein that plays an important role in the disorder. The researchers have found that disruption of the protein, Histone deacetylase 3 (HDAC3), can lead to cognitive and social impairment.
The finding, published in the journal Nature Neuroscience, could open up a new avenue of research into Rett Syndrome, as well as a potential target for treating the disorder. It may also have implications for our understanding of autism spectrum disorder and learning disabilities, according to lead author Alexi Nott, a former postdoc in the laboratory of Li-Huei Tsai at the Picower Institute for Learning and Memory at MIT.
Rett Syndrome is a rare genetic disorder of the brain that predominantly affects girls and leads to impaired motor function and intellectual abilities. Children with the disorder typically develop distinctive repetitive hand movements, such as wringing, as well as autistic traits.
The syndrome is known to be caused by a mutation in the MeCP2 gene. Previous research has shown that one effect of this mutation is to prevent the gene from interacting with the protein complex NCoR/HDAC3.
However, the precise function of the HDAC3 protein within the brain is still unclear. To investigate whether the protein plays a role in cognition, Nott and his colleagues carried out experiments in mice in which they deleted HDAC3.
They found that the loss of HDAC3 led to social and intellectual impairment and loss of motor coordination, as well as repetitive actions similar to the distinctive hand wringing seen in patients with Rett Syndrome.
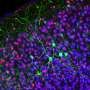
When they studied HDAC3 further, they found that the protein regulates a subset of neuronal genes, including those involved in synapse function and the transmission of nerve impulses.
The researchers then studied mice with MeCP2 deleted to model Rett Syndrome, and found that HDAC3 was unable to bind to these genes.
"So it seems that HDAC3 directly regulates these genes, and that it is dependent on MeCP2," Nott says.
To understand how the HDAC3 protein regulates the genes, the researchers then analysed them further. In particular, they hoped to identify any motifs, or patterns of DNA sequence, to which regulatory proteins, known as transcription factors, could bind. Transcription factors are proteins that control which genes are turned on or off within the genome.
They found motifs for so-called FOXO transcription factors, which were enriched in the genes that appeared to be activated by HDAC3. These FOXO transcription factors were also enriched in genes that appeared to be activated by MeCP2, according to Nott.
When HDAC3 was deleted, they found that the ability of the FOXO transcription factors was reduced, leading to a decrease in gene expression.
That made sense, as acetylation of the FOXO transcription factors is known to affect their ability to bind to DNA. Deleting HDAC3 increased the acetylation levels of the FOXO transcription factors, reducing their ability to bind to DNA.
To find out if this effect could be seen in people with Rett Syndrome, the researchers then analyzed cells from a patient with the genetic mutation thought to block the binding of HDAC3 and MeCP2.
The authors obtained the patient cells as induced pluripotent stem cells, or adult cells that have been reprogrammed into an embryonic stem cell-like state. These were then further re-programmed into neural progenitor cells.
"We found that there was decreased HDAC3 binding in the presence of this Rett mutation, and there was also decreased binding of the transcription factor, FOXO3, and this corresponded with decreased gene expression," says Nott.
When the researchers corrected the Rett mutation in the cells, using the CRISPR/Cas9 gene editing tool, they found that both HDAC3 and FOXO3 binding recovered, and the changes in gene expression were eliminated, Nott says.
The paper nicely links MeCP2, a classic DNA binding protein, to HDAC3, a histone modifying enzyme, according to Hongjun Song, a professor of neurology at Johns Hopkins University Medical School, who was not involved in the research. "This therefore links different epigenetic mechanisms together to regulate important neuronal function," he says.
"Because mutation of MeCP2 causes Rett syndrome, the current study also provides potential therapeutic treatment strategies," Song adds.
The findings should offer researchers a new avenue to explore in their search for the mechanism behind Rett Syndrome, as well as potential treatments, Nott says.
But it could also have a wider impact, since there are other core components of the HDAC3 complex that can be affected by mutations known to cause autism spectrum disorder or intellectual disability, he says.
"Because there are mutations in additional components of the complex, which have links with autism and intellectual disability, I believe that regulation of gene expression with HDAC3 might be more widely applicable, although this has yet to be tested," he says.
More information: Alexi Nott et al. Histone deacetylase 3 associates with MeCP2 to regulate FOXO and social behavior, Nature Neuroscience (2016). DOI: 10.1038/nn.4347