Model Predicts Colon Cancer Inheritable Genetic Defects
Researchers from the Johns Hopkins University and other institutions have developed a new prediction model for genetic defects known as Lynch syndrome, which predisposes families to develop colorectal cancer.
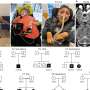
The model, called MMRpro, is based on an individual’s detailed family history of colorectal and endometrial cancer, as well as knowledge of how genetic mutations manifest themselves—in the form of tumors. It can assess a person’s probability of carrying a particular defect within so-called mismatch repair genes. The study is published in the September 27, 2006, issue of the Journal of the American Medical Association (JAMA).
“Genetic defects can be passed from parents to their children; as a result, colon cancer runs in families. Our model will help identify individuals likely to have particular genetic defects. The results will give them useful information about their colon cancer risk before they decide whether to undergo invasive screenings or expensive genetic testing,” said Sining Chen, PhD, lead author of the study and an assistant professor in the Johns Hopkins Bloomberg School of Public Health’s Department of Environmental Health Sciences.
Lynch syndrome, also known as hereditary nonpolyposis colorectal cancer, is characterized by the inheritance of defects in the MLH1, MSH2 and MSH6 genes. These three genes help repair mismatches that can occur during the duplication of the genetic code when new cells are made, and are known as mismatch repair (MMR) genes. Of the projected 600,000 MMR mutation carriers in the United States, each has approximately a 50 percent chance of being diagnosed with colorectal cancer by age 70. Women also have a 50 percent chance of developing endometrial cancer, explained Giovanni Parmigiani, PhD, senior author of the study and a professor of oncology, biostatistics and pathology at Johns Hopkins University.
The researchers applied MMRpro software to 279 individuals’ family histories. The study participants were tested with sensitive laboratory mutation-detection techniques. MMRpro predictions were compared to the laboratory test results, as well as predictions made with widely used assessment guidelines.
The researchers found that MMRpro more accurately predicted mutation carriers than two other assessment tools—the Bethesda guidelines and the Amsterdam criteria— that are currently available to families faced with the possibility that they have inherited the genetic defects related to colon cancer. MMRpro can identify more mutation carriers and fewer non-carriers than other assessment tools. MMRpro was able to access individuals who already have cancer as well as those who do not. Existing assessment tools can only be applied to individuals with cancer. In addition, certain MMR genetic mutations are hard to detect in laboratory tests. MMRpro was able to provide a useful risk assessment when conventional laboratory tests did not find a genetic mutation.
“Colorectal cancer is the second largest cause of cancer deaths in the US. It is also one of the most preventable forms of cancer. We expect that MMRpro will contribute significantly to controlling the disease by prioritizing high risk individuals for intensive screening and early detection,” said Chen. “We also expect that it will be a tool for investigators interested in understanding inherited colorectal cancer, allowing them to select families to more efficiently study these genetic defects.”
The study authors warn that MMRpro results should be interpreted by physicians and cancer counselors. The model software is available at astor.som.jhmi.edu/BayesMendel and at www8.utsouthwestern.edu/utsw/c … 829/files/65844.html .
“Prediction of Germline Mutations and Cancer Risk in the Lynch Syndrome” was written by Sining Chen, Wenyi Wang, Giovanni Parmigiani, Kenneth W. Kinzler, Francis M. Giardiello and Kathy Romans at Johns Hopkins University. Additional co-authors are Shing Lee, Khedoudja Nafa, Johanna Lee, Patrice Watson, Stephen B. Gruber, David Euhus, Jeremy Jass, Steven Gallinger, Noralane Lindor, Graham Casey, Nathan Ellis, the Colon Cancer Family Registry and Kenneth Offit.
Source: Johns Hopkins Bloomberg School of Public Health