Technique predicts hep C treatment success
By identifying genes that respond to interferon -- a drug commonly used to treat hepatitis C viral infections and certain types of cancers -- researchers have devised a novel way of predicting patient response to treatment.
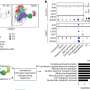
Scientists from Indiana University Bloomington, the University of Haifa (Israel) Insititue of Evolution, St. Louis University and the University of Pittsburgh used a blind, statistical approach to identify 36 genes that are not only actively expressed in the presence of interferon, but also are turned on in patients whose virus counts are dramatically diminished. The researchers describe the technique and report the results of the first test in last week's Public Library of Science (PLoS): ONE.
"This method gives us the opportunity of identifying genes that are import in the response to any drug," said IUB biologist Milton Taylor, who led the study. "This method is not necessarily confined to hepatitis C. In this case we were just using interferon and hepatitis C to see if the method works."
A growing consensus among medical scientists holds that one reason why patients respond differently -- sometimes very differently -- to a given treatment is due to the patients' unique genetic identities. But how are scientists to know which genes -- out of the 50,000 that make up the human genome -- are actually involved in mitigating, say, the flu, herpes or Hodgkin's Lymphoma?
It is not enough to simply look at what genes are turned on in the midst of an infection, or even to look at which genes are most active in patients who are faring well with a prescribed treatment, Taylor says. So he and University of Haifa mathematician Leonid Brodsky decided to delineate the most important genes by combining viral counts in the blood with gene expression across time for each individual patient.
The scientists examined expression patterns of 22,000 genes in 69 patients at six different time points during treatment. Their analysis turned up 36 candidate genes that are closely associated with virus removal in patients. A quarter of these 36 have no known function. Lending credibility to their methodology, however, a sweep of the literature shows that nearly all 36 genes have previously been identified as playing a role -- known or unknown -- in the human response to interferon treatment. Using other methods, Taylor says, a researcher would have to examine perhaps 1,000 genes altered by the treatment and from these decide by other means which were most important.
Citation: "A Novel Unsupervised Method to Identify Genes Important in the Anti-viral Response: Application to Interferon/Ribavirin in Hepatitis C Patients" PLoS: ONE, 07/04/2007
Source: Indiana University