Researchers identify proteins in lung cancer cells that may provide potential drug targets
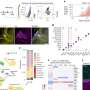
Researchers from Boston University School of Medicine (BUSM) and the Boston University Biomedical Engineering Department have identified a number of proteins whose activation allows them to distinguish between cancer and normal cells with almost 97 percent accuracy. In addition, the BU researchers have developed a new computational strategy to analyze this data and specifically identify key biological pathways (molecular circuits) that are active in cancer and "dormant" in normal cells.
The study which appears in the November 25th issue of PLoS ONE, will ultimately lead to the development of drugs specifically aimed to inhibit these proteins.
According to the BU researchers, there are many features that make cancer cells different from normal cells. They look different histologically, they proliferate and divide at different rates, they are immortal unlike normal cells, and are less communicative with their neighbor cells. They are also more "selfish" in refusing to commit suicide (programmed cell death) which normal cells do when their genomes become unstable.
Much of the cellular machinery involved with these biological processes is controlled by a command control and communication system called signal transduction. Signal transduction is in large part controlled by a process called phosphorylation. When a protein is phosphorylated it either becomes active or repressed depending on its special function. "Therefore, identifying the phosphorylation status of proteins in cancer cells versus normal cells provides us with a unique ability to understand and perhaps intervene with the command and control center of cancer cells," said co-senior author Simon Kasif, PhD, who is the co-director of the Center of Advanced Genomic Technology and a professor in the department of biomedical engineering at BU. "Drugs are most effective on cancers when they attack the proteins that are activated," he added.
While cancers are highly heterogeous in their make-up, the BU researchers believe that a drug that would target this collection of proteins would be effective treatment for most lung cancers.
"This is the first statistically validated phosphopeptide signature to diagnose any disease, much less cancer or lung cancer," explained senior co-author Martin Steffen, MD, PhD, an assistant professor of pathology and laboratory medicine at BUSM, and director, Proteomics Core Facility at BUSM.
Source: Boston University Medical Center