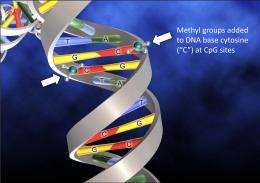
Putting the DNA back into DNA methylation. Credit: Adrian Bird
A study performed by scientists in Dirk Schübeler's team at the Friedrich Miescher Institute for Biomedical Research in Basel identifies DNA sequences that autonomously determine DNA methylation patterns. Genomic patterns of cytosine methylation are important for the proper functioning of cells during self renewal and development, and are often disrupted in disease. In a study published in the latest issue of the scientific journal Nature Genetics, Schübeler and his team show that the binding of transcription factors to specific DNA sequences determines DNA methylation levels at gene regulatory regions.
The addition of a small methyl molecule to cytosine, one of the four bases of DNA, can have far reaching consequences. It can epigenetically inactivate gene expression and thereby regulate cell fate.
In the last couple of years, scientists have analyzed DNA methylation patterns in many different situations in healthy and diseased cells. Further studies analyzed the processes and proteins that add or remove methyl groups. Despite the wealth of knowledge gained in these areas, many of the basic principles governing the patterns of DNA modifications have remain ill-defined. In the words of Dirk Schübeler, a Group leader at the FMI, "We have a fair understanding of which nucleotide is methylated at a particular stage and how the methylation is achieved, but it was unclear why one particular regulatory region was methylated and the neighboring one was not".
An analysis of the promoter regions of several genes performed by the FMI graduate student, Florian Lienert, has now identified how DNA sequence itself controls DNA methylation. "We were surprised to see that the DNA sequence of promoter regions controls its own DNA methylation fairly autonomously," said Lienert. Using gene targeting in mouse stem cells the scientists have been able to isolate small sequence modules, which they termed methylation determining regions (MDRs), that allow the recapitulation of methylation patterns. MDRs can mediate both the protection of promoters from DNA methylation as well as their methylation in a different cell type. Their activity depends on the developmental stage and the presence of motifs for the binding of regulatory proteins as well as on the base distribution in a promoter. "It is exciting to see that DNA sequence regulates this important process and to learn the basic constraints of this regulation", said Schübeler. "It predicts that sequence variations between individuals can lead to different methylation patterns. This will be relevant when analyzing and understanding methylation differences in health and disease."
More information: Lienert F, et al. (2011) Identification of genetic elements that autonomously determine DNA methylation states, Nature Genetics 43, 1091-1097, doi:10.1038/ng.946
Provided by Friedrich Miescher Institute for Biomedical Research