Chinese researchers tap GPU supercomputer for world's first simulation of complete H1N1 virus
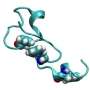
Chinese researchers achieved a major breakthrough in the race to battle influenza by using NVIDIA Tesla GPUs to create the world's first computer simulation of a whole H1N1 influenza virus at the atomic level.
Researchers at the Institute of Process Engineering of Chinese Academy of Sciences (CAS-IPE) are using molecular-dynamics simulations as a "computational microscope" to peer into the atomic structure of the H1N1 virus. Using the Mole-8.5 GPU-accelerated supercomputer, which includes more than 2,200 NVIDIA Tesla GPUs, researchers were able to simulate the whole H1N1 influenza virus, enabling them to verify current theoretical and experimental understandings of the virus.
"The Mole-8.5 GPU supercomputer is enabling us to perform scientific research that simply was not possible before," said Dr. Ying Ren, assistant professor at CAS-IPE. "This research is an important step in developing more effective ways to control epidemics and create anti-viral drugs."
Studying bacteria and viruses in laboratory experiments is difficult because reactions are often too fast and delicate to capture. And computer simulations of these systems had previously been beyond the reach of supercomputers, due to the complexity of simulating billions of particles with the right environmental conditions.
The CAS-IPE researchers made the simulation breakthrough by developing a molecular dynamics simulation application that takes advantage of GPU acceleration. It was run on the Mole-8.5 GPU supercomputer, which is comprised of 288 server nodes. The system was able to simulate 770 picoseconds per day with an integration time step of 1 femtosecond for 300 million atoms or radicals.
More information: 1. J. Xu, X. Wang, X. He, Y. Ren, W. Ge.J. Li, Application of the Mole-8.5 supercomputer: Probing the whole influenza virion at the atomic level. Chinese Science Bulletin, 2011. 56: p. 2114-2118.
2. J. Xu, Y. Ren, W. Ge, X. Yu, X. Yang.J. Li, Molecular dynamics simulation of macromolecules using graphics processing unit. Molecular Simulation, 2010. 36: p. 1131-1140.