(Medical Xpress) -- By developing a large scale gene expression map for retinal cell types, FMI Neurobiologists have been able to identify the cells in the retina, where the genes causing retinal diseases specifically act. This narrows down the search for a better understanding of the diseases and opens up new avenues for therapeutic approaches.
Most retinal diseases lead to low vision or blindness. While some of them occur spontaneously, others like retinitis pigmentosa are inherited. To date, more than 20 different retinal diseases have been identified. What almost all of them share is a daunting lack of treatment. Inherited diseases bear the advantage that the disease-causing gene can be identified: More than 200 different genes have been identified causing visual defects once they are mutated. Unfortunately this knowledge has rarely helped elucidating the molecular mechanisms leading to the disease, information that is crucial for the development of rational therapeutic approaches. Too little is known about the presence and action of these genes and proteins in the different cells of the retina.
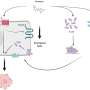
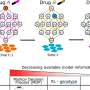
Work from the group of Botond Roska, Group leader at the Friedrich Miescher Institute for Biomedical Research, adds now a wealth of information about the expression of genes in individual retinal cells to the discussion of the pathogenesis of retinal diseases. As the FMI scientists published today online in Nature Neuroscience, they have analyzed the "transcriptome" of different cell types in the retina. The "transcriptome" describes all the genes that are being used in a cell. What they found is that even though the different retinal cells are all nerve cells working to transmit a visual signal, they still use distinct sets of genes to fulfill this function. The "transcriptome" can thus function as a barcode for the cells allowing not only a clear distinction and identification of the cells but also deduction about the mechanisms controlling their workings.
What is more, they have been able to show that disease-associated genes specifically function in a particular subset of cells in the retina. This is very valuable information because it allows narrowing down the search for a better understanding of the disease to these cells. For example, they have been able to show that in macular degeneration, where waste products of the metabolism accumulate to such a degree that the provision of the retina is impaired, the disease-causing genes are specifically expressed in the cells that should clean up in the retina, the microglia.
"We have been able to map all of the known genes associated with a retinal disease to a specific cell type in the retina and we know what other genes play a role in these cells", said Roska. "This at once opens up novel avenues for understanding the disease mechanisms and identifying therapeutic approaches for some diseases."
But all knowledge about genes and cells, and the best therapeutic approach does not suffice if the scientists cannot track the actual processing of the visual information, which means observation of the activation and deactivation of several hundred neurons in the brain region that handles visual signals, the visual cortex. A novel technological development should aid in this. Scientists from the Hungarian Academy of Sciences have developed a two photon laser microscope that allows the recording near-simultaneously from several hundred cortical neurons in vivo.
"We were the first to test and validate this new technology monitoring visual processes," said Roska. "For us it is an extremely important tool to ask basic question about visual computations and to validate our hypotheses in retinal diseases and possible therapeutic approaches."
More information: Siegert S, Cabuy E, Gross Scherf B, KohlerH, Panda S, Le Y, Fehling HJ, Gaidatzis D, Stadler MB, Roska B, 2012, Transcriptional code and disease map for adult retinal cell types. Nat Neuroscience. 2012 Jan 22. doi: 10.1038/nn.3032
Katona G, Szalay G, Maák P, Kaszás A, Veress M, Hillier D, Chiovini B, Vizi ES, Roska B, Rózsa B, 2012, Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat Methods. 2012 Jan 8. doi: 10.1038/nmeth.1851
Provided by Friedrich Miescher Institute for Biomedical Research