Setback reported in research into cancer treatment
Scientists are reporting what could be very bad news for efforts to customize cancer treatment based on each person's genes.
They have discovered big differences from place to place in the same tumor as to which genes are active or mutated. They also found differences in the genetics of the main tumor and places where the cancer has spread.
This means that the single biopsies that doctors rely on to choose drugs are probably not giving a true view of the cancer's biology. It also means that treating cancer won't be as simple as many had hoped.
By analyzing tumors in unprecedented detail, "we're finding that the deeper you go, the more you find," said one study leader, Dr. Charles Swanton of the Cancer Research UK London Research Institute in England. "It's like going from a black-and-white television with four pixels to a color television with thousands of pixels."
Yet the result is a fuzzier picture of how to treat the disease.
The study is reported in Thursday's New England Journal of Medicine.
It is a reality check for "overoptimism" in the field devoted to conquering cancer with new gene-targeting drugs, Dr. Dan Longo, a deputy editor at the journal, wrote in an editorial.
About 15 of these medicines are on the market now and hundreds more are in testing, but they have had only limited success. And the new study may help explain why.
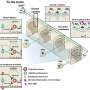
The scientists used gene sequencing to a degree that has not been done before to study primary tumors and places where they spread in four patients with advanced kidney cancer. They found that two-thirds of gene mutations they detected were not present in all areas of the same tumor. They also were stunned to see different mutations in the same gene from one part of a tumor to another.
That means a single biopsy would reveal only a minority of mutations. Still, it's not clear whether doing more biopsies would improve accuracy, or how many or how often they should be done.
Although the study involved kidney cancer, independent experts said the results should apply to other cancers such as breast, lung and colon. And previous research suggests this is so.
"This is an important paper," said Dr. Gordon Mills, co-director of the Institute for Personalized Cancer Therapy at the University of Texas MD Anderson Cancer Center.
Doctors there have been offering genetic testing to patients for several years and have a database of results on about 4,000 tumor samples. So far, about 40 percent of breast cancers have discrepancies between which genes are active in the main tumor and which ones are active where the cancer has spread, Mills said.
It costs $5,000 to $10,000 to do basic gene analysis of the main tumor, and about 10 times as much to do the kind of testing the scientists in the British study did, Mills said.
And if it were done, "we're going to find a lot of information that we don't know what to do about," such as when one biopsy suggests a certain mutation is driving the cancer and another biopsy suggests a different one is, he said.
It also takes precious time. Swanton said sequencing a patient's entire cancer genome took a very large computer four months. The amount of time required is dropping, but this type of personalized analysis is still years away from being available in the clinic, he said.
Yet the study shows that the single biopsy - "the cornerstone of personalized-medicine decisions" - is not enough, Longo wrote. And "the simple view of directing therapy on the basis of genetic tumor markers is probably too simple."
©2012 The Associated Press. All rights reserved. This material may not be published, broadcast, rewritten or redistributed.