Uncovering the genome's regulatory code
Since the sequencing of the human genome in 2001, all our genes – around 20,000 in total – have been identified. But much is still unknown – for instance where and when each is active. Next to each gene sits a short DNA segment, and the activity of this regulatory segment determines whether the gene will be turned on, where and how strongly. These short regulatory segments are as – if not more – important than the genes, themselves. Indeed, 90% of the mutations that cause disease occur in these regulatory areas. They are responsible for the proper development of tissues and organs, determining, for instance, that eye cells – and only eye cells – contain light receptors, while only pancreatic cells function to produce insulin. Clearly, a deeper understanding of this regulatory system – its mechanisms and possibilities for malfunction – may lead to advances in biomedical research, especially in developing targeted therapies for individual patients.
In spite of their importance, the "regulatory code" is not well understood. To address this problem, a research team led by Dr. Ido Amit of the Weizmann Institute Immunology Department, together with scientists from the Broad Institute in Massachusetts, including Manuel Garber, Nir Yosef and Aviv Regev, and Nir Friedman of the Hebrew University of Jerusalem, developed an advanced, automated system for mapping these sites, and then used this system to uncover important principles how these regulatory elements function. Among other things, their study, which appeared in Molecular Cell, revealed a hierarchical structure for the regulatory code. By mapping a large number of regulatory factors, the team succeeded in revealing an overall plan for gene regulation as well as the intimate details of the mechanisms involved in the immune response.
"We are seeing a race to map the regulatory code and uncover its ties to disease and human variation that is reminiscent of the race to sequence the human genome," says Amit. "But until now, participants have faced a serious hurdle: The process used for the past 30 years to map regulatory elements has been complicated, complex and labor-intensive, requiring huge scientific consortiums. With the new method, just a handful of researchers were able to conduct a study on a similar scale to the mega-team ones, and in a fraction of the time."
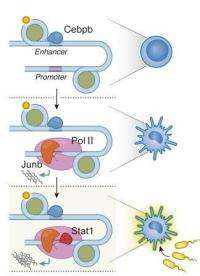
Their highly efficient, automated method enabled Amit and his team to measure a large number of regulatory proteins and their binding sites in parallel. They exposed immune cells to bacteria – setting the stage for gene activation – and then traced the actions of several dozen different regulatory proteins known to play a role in the immune response over four points in time. Not only were the researchers able to identify the binding locations of each and the genes they activate, but the levels of activation and the mechanisms employed.
One of their more significant findings was that the actions of these regulatory factors can be neatly classified into three levels in a sort of regulatory hierarchy. In the bottom tier are those factors that create the rough divisions into main cell types by directing cell differentiation. These factors are the "basic identity" guides that can, on their own, determine whether a cell will have the characteristics of a muscle cell, a nerve cell, etc. On the second tier are the regulatory factors that determine a cell's sub-identity, which they do by controlling the strength of a gene's expression. These factors are in charge of producing closely-related sub-types, for instance, muscle fibers that are either smooth or striated, or closely-related immune cells. Regulatory factors in the third tier are even more specialized: They only affect the expression of certain genes that are called into action in response to signals from outside the cell: bacterial invaders, hormones, hunger pangs, etc.
The hope is that understanding the ins and outs of the regulatory code will help researchers to understand and predict how diseases arise and progress due to malfunctions in regulatory mechanisms. In the future, understanding the regulatory program may lead to advances in rehabilitative medicine. Regulatory mechanisms could be used to redirect the differentiation of a patient's cells, which could then be reimplanted, thus avoiding the problems inherent in using donor cells.
Amit: "The new method for mapping the gene's regulatory plan may open new vistas for investigating all sorts of biological processes, including the system failures that occur in disease."
More information: www.sciencedirect.com/science/ … ii/S1097276512006570

















