August 28, 2013 feature
Keep your cool: Isothermal PCR for next-generation sequencing

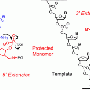
(Medical Xpress)—In the realm of DNA sequencing, the Polymerase Chain Reaction (PCR) is a well-established tool for amplifying a specific DNA sequence by reiteratively processing what's known as a reaction cocktail between different temperatures. That said, however, there are alternative methods of sequence-specific DNA amplification that do not require the reaction temperature to vary, and are therefore referred to as sequence-specific isothermal amplification techniques. Such approaches have the dual advantage of being extremely fast and not needing thermocyclers. Recently, scientists in the Genetic Systems group at Life Technologies Inc. in California developed a cost-effective in situ (literally, situated in the original, natural, or existing place) isothermal amplification method based on a so-called template walking mechanism – a tool for creating a DNA copy of the genetic information to be amplified – using a pair of low-melting temperature (Tm) solid-surface homopolymer primers and a low-Tm solution phase primers that can generate billions of monoclonal colonies in less than 30 minutes. The method can generate more than one billion submicrometer-sized colonies in a single lane of a next-generation sequencing flowchip. The researchers say that their method – Intended for next-generation sequencing (NGS), in which high-throughput technologies parallelize and thereby shorten the sequencing process – can simplify clinical genomic applications and make the long-sought $1,000 genome possible. Moreover, the team also demonstrated an alternative paired-end sequencing method that uses interstrand DNA photo cross-linking to covalently link the complementary strands of the original templates to the solid surface.
Genetic Systems Principal Scientist Kaiqin Lao discussed the research he, Zhaochun Ma and their colleagues conducted with Medical Xpress. "There are two keys to achieving cost-effective, efficient in situ PCR amplification for next-generation sequencing," Lao tells Medical Xpress. "First; the sequences of primers need to be low complexity, such as homo-polymers. Second, the distance between the two surface primers has to be within the length of the primer." In this case, Lao notes, that length is 2-4 nanometers.
One of the difficulties in working with a template walking mechanism is to find a polymerase with strong strand displacement activity, high processivity, and optimal activity around 60?C. "The melting temperature of homopolymer primers has to be lower than the reaction temperature of roughly 60?C," Lao notes, "to allow efficient primer invasion at both ends of the DNA duplex without fully denatured templates."
Another challenge was to minimize mishybridization between primers and the poly-T or TTG repeat region of the DNA insert by converting the fragment libraries to nicked libraries using a forward primer containing deoxyuridine and a reverse primer, followed by uracil-specific excision reagent treatment. "Our original protocol had chemical or thermal denaturation steps," Lao explains. ". Later, we realized that CTT triplet repeat sequences are one of the common short tandem repeats in more complex genomes that solution primer can anneal and extend to generate shorter templates. To overcome this, we used the reagent treatment to generate nicked overhang poly-T to prime with surface primer while the genomic sequences remained duplex in order to prevent solution primer to anneal." The researchers nicknamed this strategy Watson protects Crick, Lao quips.
Lao points out that they also developed an alternative paired-end sequencing method that differs from bPCR (bridging, or jumping, PCR) using interstrand DNA photo cross-linking to covalently link the complementary strands of the original templates to the solid surface. "Template walking has one solid phase primer and one solution phase primer, so when complete only one end of the colonies is anchored to the solid surface," Lao explains. "For pair-end reads, the other end needs to be attached to the solid surface before reverse sequencing. We spent quite some time searching for efficient DNA cross-linking reagents before we indentified the CNVK-base as a fast photo cross linking reagent. We also spent a great amount of time to find a specific buffer composition that doesn't; react with the CNVK-base during forward sequencing, and at the same time protects templates from photo-damage."
The scientists addressed these complex challenges thanks to a single insight – namely, George Church's 1999 Polony paper1. "This paper has been in my mind constantly," Lao acknowledges. "The challenge is how to immobilize amplified templates while allowing the walking strands to diffuse to nearby colonies to form polyclonal colonies."
The scientists state that their method can be used on NGS platforms to make the $1,000 genome possible by pumping in fresh enzyme mixtures. "In contrast," Lao notes, "bPCR reagent consumption will be 17 or 35 times more than with template walking if we assume that the same amount of enzymes was used for each cycle. Furthermore, bPCR has both primers on the surface to form both strands, which need to remove one of the complementary strands before the sequencing. 'In contrast, all extended surface primers have sequenceable templates, so that the colony intensity is at least two times higher than bPCR, which will allow longer reads and simpler instruments with less expensive lasers and imaging systems.
"Template walking has significantly reduced the colony formation reagents cost by over 10-20 times," Lao continues, "which is about 60% of total expense of current NGS sequencing reagents based on bPCR, while the sequencing primers and probes are less than 40%. From a purely technical perspective, $1,000 genomic sequencing has been achieved with today's technologies based on the cost of goods."
Looking ahead, Lao says that template walking can also be used to replace general PCR, so it is possible that it could be implemented on any analogue or digital PCR platforms. "This will also enable battery powered handheld devices for point-of-care diagnostic applications." In addition, he adds, template walking will make NGS as simple as quantitative PCR (qPCR) with integrated samples-to-library-to-colonies without thermal cycling – which is currently one of the main challenges in lab-on-chip technology.
In terms of other areas of research that might benefit from their study, Lao concludes that template walking's isothermal amplification nature may make it possible to enable in vivo amplification in whole, or even living, cells.
More information: Isothermal amplification method for next-generation sequencing, PNAS Published online before print August 12, 2013, doi:10.1073/pnas.1311334110
References
1In situ localized amplification and contact replication of many individual DNA molecules, Nucleic Acids Research 27(24):e34 (PDF)
© 2013 Medical Xpress. All rights reserved.