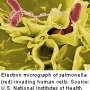
Researchers track antibiotic-resistant strains of Salmonella from farm to fork
Continuing research on Salmonella may enable researchers to identify and track strains of antibiotic resistant bacteria as they evolve and spread, according to researchers in Penn State's College of Agricultural Sciences.
Tracing the transmission of individual strains from agricultural environments to humans through the food system is difficult because of the rapid evolution of resistance patterns in these bacteria. Resistance patterns change so quickly that, until now, it has been impossible to determine where some highly resistant strains are coming from.
Michael DiMarzio, a doctoral candidate in food science working under the direction of Edward Dudley, associate professor and Casida Development Professor of Food Science, developed a method for identifying and tracking strains of Salmonella enterica serological variant Typhimurium as they evolve and spread.
Every year in the United States, the various strains of Salmonella together are responsible for an estimated 1 million illnesses, 20,000 hospitalizations and 400 deaths at an economic cost exceeding $3 billion. Salmonella Typhimurium accounts for at least 15 percent of clinically reported salmonellosis infections in humans nationally. The number of antibiotic-resistant isolates identified in humans is increasing steadily, suggesting that the spread of antibiotic-resistant strains is a major threat to public health.
"Typhimurium infections have exhibited a gradual decline in susceptibility to traditional antibiotics, a trend that is concerning in light of this pathogen's broad host range and its potential to spread antibiotic resistance determinants to other bacteria," DiMarzio said. "Now more than ever, it is imperative to effectively monitor the transmission of Salmonella Typhimurium throughout the food system to implement effective control measures."
Building on recent research done in Dudley's lab, DiMarzio developed the new approach to identify antibiotic resistant strains of Salmonella Typhimurium focusing on virulence genes and novel regions of the bacteria's DNA known as clustered regularly interspaced short palindromic repeats, or CRISPRs. They report their results in the September issue of Antimicrobial Agents and Chemotherapy.
CRISPRs are present in many foodborne pathogens. The researchers demonstrated that CRISPR sequences can be used to identify populations of Salmonella with common antibiotic-resistance patterns in both animals and humans.
"Specifically, we were able to use CRISPRs to separate isolates by their propensity for resistance to seven common veterinary and human clinical antibiotics," DiMarzio said. "Our research demonstrates that CRISPRs are a novel tool for tracing the transmission of antibiotic-resistant Salmonella Typhimurium from farm to fork."
DiMarzio found that several subtypes of Salmonella Typhimurium showed up repeatedly in the frozen collection of Salmonella samples taken from cows, pigs and chickens in Penn State's Animal Diagnostic Laboratory. In this case, researchers looked at 84 unique Salmonella Typhimurium isolates collected from 2008 to 2011.
"We know those strains are widely disbursed, and the thing they have in common is that they have noticeably higher levels of antibiotic resistance," he said. "So we examined clinical samples of Salmonella taken from humans, and it turned out that we see an overlap—the ones we see in humans are the ones we see a lot in animals. You would expect that, but it is confirmation that our method works."
DiMarzio noted that the researchers identified subsets of the overall Salmonella bacteria population that seem to be more prone to acquiring antibiotic resistance.
"Our challenge now is to learn what makes those strains different—why do some strains acquire resistance while others don't, even though both are circulating widely among animal populations?" he said. "We will need to know that to try to control them."