New statistical tools being developed for mining cancer data
Researchers at Rice University, Baylor College of Medicine (BCM) and the University of Texas at Austin are working together to create new statistical tools that can find clues about cancer that are hidden like needles in enormous haystacks of raw data.
"The motivation for this is all of these new high-throughput medical technologies that allow clinicians to produce tons of molecular data about cancer," said project lead Genevera Allen, a statistician with joint appointments at Rice and BCM. "For example, when a tumor is removed from a cancer patient, researchers can conduct genomic, proteomic and metabolomic scans that measure nearly every possible aspect of the tumor, including the number and location of genetic mutations and which genes are turned off and on. The end result is that for one tumor, you can have measurements on millions of variables."
This type of data exists—the National Institutes of Health (NIH) has compiled such profiles for thousands of cancer patients—but scientists don't yet have a way to use the data to defeat cancer.
Allen and her collaborators hope to change that, thanks to a new $1.3 million federal grant that will allow them to create a new statistical framework for integrated analysis of multiple sets of high-dimensional data measured on the same group of subjects.
"There are a couple of things that make this challenging," said Allen, the principal investigator (PI) on the new grant, which was awarded jointly by the National Science Foundation and the NIH. "First, the data produced by these high-throughput technologies can be very different, so much so that you get into apples-to-oranges problems when you try in make comparisons. Second, for scientists to leverage all of this data and better understand the molecular basis of cancer, these varied 'omics' data sets need to be combined into a single multivariate statistical model."
For example, Allen said, some tests, like gene-expression microarrays and methylation arrays, return "continuous data," numbers with decimal places that represent the amounts of a particular protein or biomarker. Other tests, like RNA-sequencing, return "count data," integers that indicate how often a biomarker shows up. And for yet other tests, the output is "binary data." An example of this would be a test for a specific mutation that produces a zero if the mutation does not occur and a one if it does.
"Right now, the state of the art for analyzing these millions of biomarkers would be to create one data matrix—think one Excel spreadsheet—where all the numbers are continuous and can be represented with bell-shaped curves," said Allen, Rice's Dobelman Family Junior Chair of Statistics and assistant professor of statistics and electrical and computer engineering. "That's very limiting for two reasons. First, for all noncontinuous variables—like the binary value related to a specific mutation—this isn't useful. Second, we don't want to just analyze the mutation status by itself. It's likely that the mutation affects a bunch of these other variables, like epigenetic markers and which genes are turned on and off. Cancer is complex. It's the result of many things coming together in a particular way. Why should we analyze each of these variables separately when we've got all of this data?"
Developing a framework where continuous and noncontinuous variables can be analyzed simultaneously won't be easy. For starters, most of the techniques that statisticians have developed for parallel analysis of three or more variables—a process called multivariate analysis—only work for continuous data.
"It is a multivariate problem, and that's how we're approaching it," Allen said. "But a proper multivariate distribution does not exist for this, so we have to create one mathematically."
To do this, Allen and her collaborators—co-PIs Zhandong Liu of BCM and Pradeep Ravikumar of UT Austin—are creating a mathematical framework that will allow them to find the "conditional dependence relationships" between any two variables.
To illustrate how conditional dependence works, Allen suggested considering three variables related to childhood growth—age, IQ and shoe size. In a typical child, all three increase together.
"If we looked at a large dataset, we would see a relationship between IQ and shoe size," she said. "In reality, there's no direct relationship between shoe size and IQ. They happen to go up at the same time, but in reality, each of them is conditionally dependent upon age."
For cancer genes, where the relationships aren't as obvious, developing a mathematical technique to decipher conditional dependence could avoid the need to rule out such errors through years of expensive and time-consuming biological experiments.
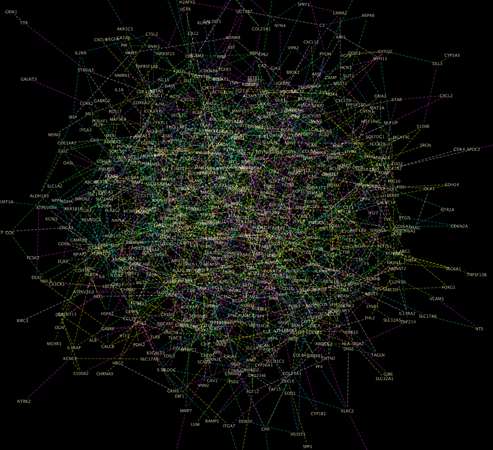
Allen and her collaborators have already illustrated how to use the technique. They've produced a network model for a half-million biomarkers related to a type of brain cancer called glioblastoma. The model acts as a sort of road map to guide researchers to the relationships that are most important in the data.
"All these lines tell us which genetic biomarkers are conditionally dependent upon one another," she said in reference to the myriad connections in the model. "These were all determined mathematically, but our collaborators will test some of these relationships experimentally and confirm that the connections exist."
Allen said the team's technique will also be useful for big data challenges that exist in fields ranging from retail marketing to national security.
"This is a very general mathematical framework," she said. "That's why I do math. It works for everything."