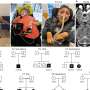
Population structure and genetic interactions of Streptococcus pneumoniae.(a) Family tree of a pneumococcal population with connections between recipients and potential donors of DNA fragments. (b) Most prevalent forms of the bacterium detected in the Maela population. [doi:10.1038/ng.2895]
Researchers have found that a harmless bacterium could be a source for antibiotic resistance in treatments against such diseases as pneumonia, bacteraemia and meningitis. Their results could help in the design of better strategies to deal with the emergence of antibiotic resistance.
Streptococcus pneumoniae is a bacterium that is a major global health problem. Although there are vaccines currently available against this bacterium, S. pneumoniae can evade the vaccine by exchanging its DNA in a process known as recombination. This can include the gain of antibiotic-resistant genetic variants and increase the risk of wider spread of antibiotic resistance.
In this study, a seemingly harmless strain of the bacterium, known as non-typable or NT, was the most common type of S. pneumoniae found in over 3000 samples from individuals in a refugee camp. Although harmless, this strain is exchanging DNA at a faster rate than the other more harmful forms of the bacterium, including DNA carrying genes that carry antibiotic resistance.
"Vaccination programs have had a very positive impact on the rates of infection with resistant S. pneumoniae but we must remain vigilant as this pathogen is a master of adaptation with the potential to overcome our attempts at controlling it," says Professor Stephen Bentley, senior author from the Wellcome Trust Sanger Institute. "This study shows that if we want to effectively deal with resistance, we have to keep track of this form of S. pneumoniae - we simply cannot afford to ignore it."
The team sequenced over 3000 samples of Streptococcus pneumoniae from almost 1000 infants and mothers from a refugee camp on the border between Myanmar and Thailand, to understand the evolution of the bacterium during healthy carriage and transmission from person to person. The proportion of people who carry S. pneumoniae tends to be greater in developing countries, as are the rates of pneumococcal disease.
They found that the most common form of this bacterium was one that rarely causes disease. These NT pneumococci lack the sugar capsule that generally coats all other disease-causing forms of the bacterium. However, the lack of a capsule means that the NT pneumococci can exchange DNA more readily than those enclosed in a sugar capsule.
The team found DNA exchange was exceptionally high in regions of the genome that are associated with antibiotic-resistance. They found that the swapping of antibiotic resistant regions of the genome has mediated the spread of resistance to the major family of ß-Lactam antibiotics which includes penicillin. This is consistent with the overuse of ß-Lactam antibiotics in the region since the 1990s.
"We can see with greater resolution, the process of recombination that provides an enhanced understanding of how this bacterium might acquire and lose antibiotic resistance," says Claire Chewapreecha, first author from the Wellcome Trust Sanger Institute. "The reservoir of antibiotic-resistant genes is a real worry because it gives the pathogen population the potential to adapt to antibiotics more rapidly."
Current vaccines target the sugar capsules that enclose these harmful bacteria. As the NT pneumococci does not have the sugar capsule, current vaccine cannot protect against the possibility of resistance genes being transferred from NT pneumococci.
"These NT pneumococci without the capsule are a real issue for us in the battle against antibiotic resistance," says Dr Paul Turner, author from the Shoklo Malaria Research Unit. "The two problems with NT pneumococci are firstly the prevalence of DNA exchange, especially antibiotic resistant genetic material, and secondly current vaccines cannot protect us against it - this is a dangerous combination. This research will help teams develop future intervention strategies."
More information: "Dense genomic sampling identifies highways of pneumococcal recombination." Claire Chewapreecha, et al. Nature Genetics 2014. DOI: 10.1038/ng.2895
Journal information: Nature Genetics
Provided by Wellcome Trust Sanger Institute
![Population structure and genetic interactions of Streptococcus pneumoniae.(a) Family tree of a pneumococcal population with connections between recipients and potential donors of DNA fragments. (b) Most prevalent forms of the bacterium detected in the Maela population. [doi:10.1038/ng.2895] Exchange of genetic material between harmless bacteria could be reservoir of antibiotic resistance](https://scx1.b-cdn.net/csz/news/800a/2014/exchangeofge.jpg)