IBM research breakthrough helps public health officials improve food safety
IBM today announced a first-of-a-kind system that is designed to help food retailers, distributors and public health officials predict the most likely contaminated food sources and accelerate the investigation of food-borne disease outbreaks. Using novel algorithms, visualization, and statistical techniques, the tool can use information on the date and location of billions of supermarket food items sold each week to quickly identify with high probability a set of potentially "guilty" products with in as few as 10 outbreak case reports. This research was published today in the peer-reviewed journal PLOS Computational Biology together with collaborators from Johns Hopkins University, Purdue University and the German Federal Institute for Risk Assessment (BfR).
Food-borne disease outbreaks of recent years demonstrate that due to increasingly interconnected supply chains food-related crisis situations have the potential to affect thousands of people, leading to significant healthcare costs, loss of revenue for food companies, and —in the worst cases— death. In the United States alone, one in six people are affected by food-borne diseases each year, resulting in 128,000 hospitalizations, 3,000 deaths, and a nearly $80B economic burden.
When a food-borne disease outbreak is detected, identifying the contaminated food quickly is vital to minimize the spread of illness and limit economic losses. However, the time required to detect it may range from days to weeks, creating extensive strain on the public health system.
Perhaps surprisingly, the petabytes of retail sales data have never before been used to accelerate the identification of contaminated food. In fact, this data already exists as part of the inventory systems used by retailers and distributors today, which manage up to 30,000 food items at any given time with nearly 3,000 of them being perishable.
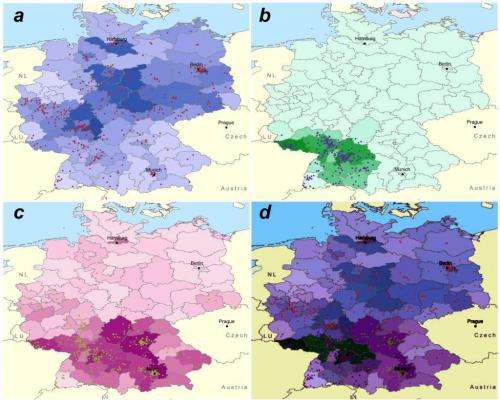
Recognizing this issue, IBM scientists built a system that automatically identifies, contextualizes and displays data from multiple sources to help reduce the time to identify the mostly likely contaminated sources by a factor of days or weeks. It integrates pre-computed retail data with geocoded public health data to allow investigators to see the distribution of suspect foods and, selecting an area of the map, view public health case reports and lab reports from clinical encounters. The algorithm effectively learns from every new report and re-calculates the probability of each food that might be causing the illness.
"Predictive analytics based on location, content, and context are driving our ability to quickly discover hidden patterns and relationships from diverse public health and retail data," said James Kaufman, Manager of Public Health Research for IBM Research, "We are working with our public health clients and with retailers in the U.S. to scale this research prototype and begin focusing on the 1.7B supermarket items sold each week in the United States."
How it Works
To demonstrate the system's effectiveness, IBM scientists worked with the Department of Biological Safety of the German Federal Institute for Risk Assessment. In this demonstration, the scientists simulated 60,000 outbreaks of food-borne disease across 600 products using real-world food sales data from Germany.
Unfortunately, in real life cases of food-borne disease do not show up all at once as outbreaks are reported over a period of time. Depending on the circumstances, it takes public health officials weeks or months to identify the real cause, sometimes this is even not possible at all. If the relevant data would be provided by the retail companies this could be improved significantly already people were sick.
The success of an outbreak investigation often depends on the willingness of private sector stakeholders to collaborate pro-actively with public health officials. This research illustrates an approach to create significant improvements without the need for any regulatory changes. This can be achieved by combining innovative software technology with already existing data and the willingness to share this information in crisis situations between private and public sector organizations," said Dr. Bernd Appel, Head of the Department Biological Safety, BfR.