Structure of the Aurora A kinase protein. Based on PyMOL molecular visualization system rendering of Protein Data Bank (PDB) 1mq4. Credit: Created by Emw, Wikimedia CC BY-SA 3.0.
(Medical Xpress)—In the ongoing quest to design new beneficial molecules or identify potential drugs catalogued in pharmaceutically databases, a critical requirement is determining how a ligand (typically a modulator, or signal-triggering molecule) binds to a therapeutic protein. Currently, most drug design protocols the most potent ligand is chosen – but at the cost of diminished target protein specificity. On the other hand, such small-molecule ligands having what is known as a high TC50 value, meaning that a larger amount of the drug is needed to be effective – and they may therefore be rejected. Recently, however, scientists at Jawaharlal Nehru Centre for Advanced Scientific Research, India combined surface-enhanced Raman spectroscopy (SERS) and molecular dynamics (MD) simulation to identify the precise location on a target protein where modulators bind. (SERS is a surface-sensitive technique used to analyze nanoscale composition that enhances normal Raman signals by up to eight orders of magnitude; MD simulates physical movements of atoms and molecules to refine three-dimensional structures of proteins and other macromolecules.)
In addition, the researchers demonstrated the new tool's efficacy by determining the selective binding of the antihypertension drug felodipine to the oncogenic (that is, causing development of a tumor or tumors) Aurora A kinase human enzyme. Moreover, the researchers employed SERS to selectively inhibit Aurora A's antihypertensive property while retaining its anti-cancer property – to their knowledge for the first time – and corroborated this selective inhibition using molecular docking studies and MD simulations. (Molecular docking is a molecular modeling method that predicts the preferred orientation of two bound molecules, providing the ability to predict molecular binding affinity.) The scientists therefore conclude that their ability to acquire a priori knowledge of protein structure indicates that their new protocol has shown very strong potential for providing advances in drug discovery – especially the development of new higher-potency medications.
Prof. Chandrabhas Narayana discussed the paper that he and his co-authors published in Proceedings of the National Academy of Sciences with Medical Xpress. Regarding the use of SERS to identify the binding site of small molecules on a therapeutically important protein, Narayana notes that SERS has traditionally been used to detect small molecules having high Raman scattering cross sections – that is, the distortion of a molecule in an electric field, which is a function of the degree to which it can be polarized. "However," Narayana tells Medical Xpress, "using SERS to detect large molecules like proteins, and subsequently interpreting the data, are challenging tasks that are not generally explored. Therefore, little or no structural information is available for most of the therapeutically important proteins. We're the first group to use SERS to investigate these proteins."
That said, Narayana points out that "the biggest challenge was getting SERS measurement of the proteins, where surface charge and surface hydrophobicity determine the nanoparticle protein adsorption – and standard Raman spectroscopy is difficult to perform on these therapeutically important proteins, due largely to the possibility of the nanoparticles affecting protein activities. Another challenge," he continues, "was acquiring protein structural information, since MD simulation and AutoDock seldom predict the correct structure or the small molecule binding of such systems." AutoDock is a suite of automated molecular docking tools designed to predict how small molecules bind to a receptor of known 3D structure.
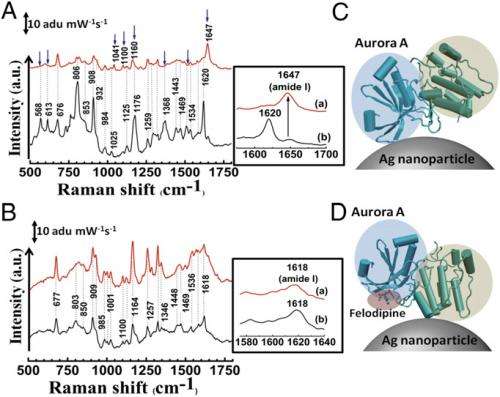
SERS study of specific binding of felodipine to Aurora A. (A) SERS spectrum of Aurora A (black) and Aurora A complexed with felodipine (red). (B) SERS spectrum of Aurora B (black) and after treatment with felodipine (red). The change in position of modes and appearance of new modes are indicated by blue arrows in A and amide I bands are highlighted in Insets. (C) Mode of attachment of Aurora A to a silver nanoparticle and (D) change in orientation of Aurora A on the silver nanoparticle surface on complexation with felodipine. The N-terminal β-sheet–rich domain and the C-terminal α-helix–rich domains are highlighted in blue and green, respectively. The bound felodipine in D is highlighted in red. Credit: Copyright © PNAS, doi:10.1073/pnas.1402695111
Narayana adds that yet another challenge was to understand how the protein molecule attaches to the surface of the noble metal nanoparticles, the latter being responsible for the SERS phenomenon. (A noble metal – examples include gold, silver and platinum – is resistant to chemical action, corrosion and acids.) "The most important challenge is that a surface-enhanced Raman spectroscopic measurement of protein is not a complete Raman spectrum," he explains, "but as its name indicates focuses close to the nanoparticle surface. Therefore, the interpretations are not very straightforward. Nevertheless, before conducting the SERS experiments, the docking studies resulted in very different sites – a common problem in docking studies due to multiple possible ground states. Therefore, mutation studies on these sites never gave the desired results." By utilizing SERS, molecular docking and simulation, however, the scientists were able to identify the binding sites of small molecules on proteins.
An important result was that the scientists achieved inhibition using a unique surface-binding mode. "The surface-binding mode is a previously unexplored mode of Aurora A inhibition," Narayana tells Medical Xpress. "The fact that inhibition happens when binding happens away from the active site is not only interesting, but in addition provides an alternative mode of inhibition." The researchers understood the mechanism of this inhibition – which indicated changes in the Aurora A ATP binding pocket – from their molecular dynamics simulations.
Narayana notes that molecular docking also played an important role in their research, specifically in searching the accurate hydrophobic pockets where the felodipine molecule could bind. "This technique – based on the Lamarckian genetic algorithm – saves a great deal of computational time, and has been used extensively for determining binding of small molecules to proteins. (AutoDock uses the Lamarckian genetic algorithm as one of several search algorithms to find optimal conformation with the lowest binding energy.) "The docked position forms an initial point for the molecular dynamics simulations where protein flexibility is taken into account, and provides information about the structural changes that occurred during small molecule binding to the protein."
Molecular dynamics simulation of a large molecule like protein was made possible due to the availability of fast and efficient computers, he adds. "We collaborated with the group of Prof. Hans Agren at KTH, Sweden, and were able to use their computational facilities. The molecular dynamics simulations gave us insights into the mechanism of inhibition of Aurora A on binding of felodipine through subtle variation in its structure. It also helped us to validate the docked positions of felodipine. The important thing is that we have used the code where we can look at protein as quantum mechanical in the region of interest and coarse grain elsewhere – but again, this would not have been possible without structural information and the use of SERS to verify the effects seen."
In their paper, the scientists say that felodipine-related compounds are being considered as antineoplastic therapeutics. "One of the things one should keep in mind is that as an antihypertension drug, we had to first derivatize felodipine such that it lost its hypertension control, but retained its anti-cancer property," Narayana points out. "The major challenge here is achieving specificity against the target of interest with the least possible off-targeting potential." (Derivatization is a technique used in chemistry which transforms a chemical compound into a similar chemical structure called a derivative.) "The major hurdle in using this class of compounds is to achieve optimal therapeutic concentrations to treat cancer without blocking the calcium channels essential to maintaining calcium ion homeostasis." In other words, achieving a higher therapeutic index is one of the important factors that need to be considered when designing small molecules for a specific pathological situation.
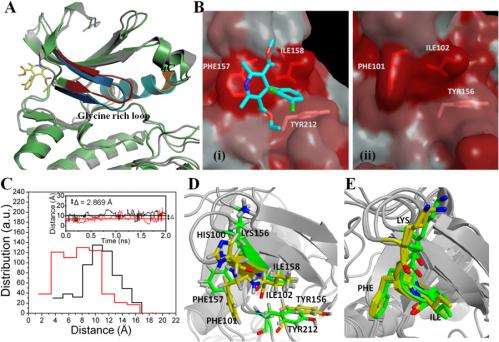
(A) Superimposed structures of Aurora A after 2 ns of MD simulation with (gray) and without (green) felodipine. The glycine-rich loop region undergoes change in conformation (colored in red and blue to denote free and felodipine-bound, respectively). The αC region is colored in orange (free) and cyan (felodipine-bound). (B) Binding region of felodipine to Aurora A (i) and the corresponding site in Aurora B (ii) represented by Eisenberg’s hydrophobicity index. The felodipine bound to Aurora A is shown in stick representation. (C) rmsd distances between center of masses of residues Phe-157 and Ile-158 (Aurora A is black) and Phe-101 and Ile-102 (Aurora B is red) obtained in 400 frames of a 2-ns MD simulation plotted as a histogram. Inset shows the variation in the rmsd distance between center of masses of Phe-157 and Ile-158 (Aurora A, in black) and Phe-101 and Ile-102 (Aurora B, in red) as a function of time. The mean averages of these distances are shown as solid lines. The difference in mean average was 2.869 Å. (D) Superimposed felodipine-binding pocket of 1-ns equilibrated structures of human Aurora A and the corresponding pocket in human Aurora B. Key residues are shown in sticks. (E) Superimposed crystal structures of human Aurora A (PDB ID codes 1MQ4 and 2WTV) and Xenopus Aurora B (PDB ID codes 2VGO and 2BFX). The carbon atoms in Aurora A and B are colored in green and yellow, respectively. Nitrogen, oxygen, and hydrogen atoms are colored in blue, red and white, respectively in D and E. Credit: Copyright © PNAS, doi:10.1073/pnas.1402695111
Several key insights and innovations were central to solving these myriad challenges. "Without co-author Prof. Tapas Kundu's discovery of felodipine being selective for Aurora kinase, as well as his identifying the biochemical components of the drug's interaction with Aurora A, our project would not have taken off," Narayana tells Medical Xpress. "In addition, due to our experience of working with proteins such as p300 and CARM1, we realized that while very interesting interpretations were possible with SERS, the lack of the protein's structure made it very difficult to take it further. Therefore, finding a protein whose structure was known gave us the first insight that this would be a good case for taking SERS beyond what we had achieved up to that point. The icing on the cake was the fact that the beta sheet and alpha helix were well separated." (The beta sheet and alpha helix (or β-sheet and α-helix, respectively) are secondary protein structures – three-dimensional forms of local segments of biopolymers such as proteins and nucleic acids.) "Over the years we've fine-tuned our SERS techniques to get the best signals without the nanoparticle effecting protein activities or protein adsorption on the nanoparticle surface. As a result, we've developed the right nanoparticle-to-protein concentration, compaction of the nanoparticle suspension to create hot spots, and retaining the pH around the protein so that it cannot denature."
Narayana notes that a properly-developed SERS methodology for individual proteins can be used to rapidly screen possible ligands, thereby leading to a range of therapeutic applications. "SERS carried out on felodipine, its derivatives, and generic drugs demonstrated that none of the derivatives and generic drugs which disturb molecular hydrophobic-to-hydrophilic distance, size, or steric hindrance in work on Aurora kinase," Narayana illustrates, "and in fact doesn't show any changes similar to felodipine." (Steric hindrance occurs when the large size of groups within a molecule prevents chemical reactions that are observed in related molecules with smaller groups.) "This tells us that if we look at the specific changes in the protein upon introducing the drug, the first test is to study its inhibition or activation properties. For this to be successful, we need to completely understand SERS binding."
Moving forward, the scientists are focused on two major steps: using this technique to in drug discovery, as well as to study other challenging problems in protein biology where structural changes would throw light on protein activities.
• Drug design/discovery:
- High-throughput procedure for rapid screening
- Optimization of platforms to investigate protein target arrays; appropriate capping agents over the SERS active nanoparticles; and nanoparticle functionalization (that is, using surface coatings to determine nanoparticle properties) to achieve control in binding at specific protein regions
• Protein Structural studies:
- Facilitating human proteome research
- Investigating protein structural changes driven by positively charged ions, or cations – for example, genetic transcription coactivation or restriction enzymes (proteins that respectively increase or decrease gene expression by binding to an activator or transcription factor
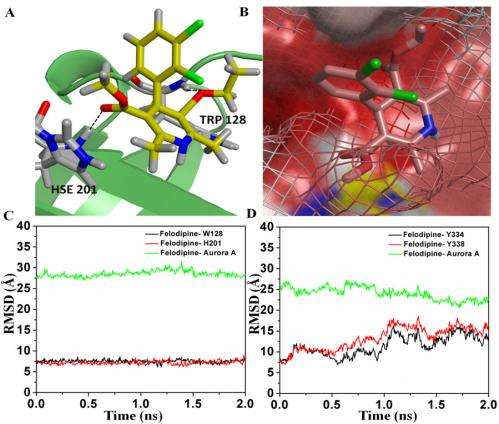
(A) The bound configuration of felodipine to Aurora A to the second binding site. The residues colored in gray are in hydrophobic interaction with felodipine. Felodipine is hydrogen-bonded to Aurora A through residues Trp-128 and His-201 with hydrogen bond distances of 2.1 and 1.9 Å, respectively (represented by dashed line). The carbon, oxygen, nitrogen, and chlorine atoms are colored in yellow, red, blue, and green, respectively. (B) The surface-binding mode of felodipine to the second binding site. The Aurora B surface is shown in wireframe to highlight the difference in the hydrophobic binding site. (C) rmsd distances between felodipine and residues Trp-128 (black) and His-201 (red) and Aurora A (green) for the second binding site. (D) rmsd distances between felodipine and residues Tyr-334 (black) and Tyr-338 (red) and Aurora A (green) for the third site. Credit: Copyright © PNAS, doi:10.1073/pnas.1402695111
Narayana also identifies several other innovations that the researchers might consider developing – specifically, a deeper understanding of tip-enhanced Raman spectroscopy (TERS) – a scanning probe microscopy technique combining the capabilities of Raman spectroscopy with the spatial resolution of an atomic force microscopy (<30 nm); in vivo application of SERS active nanoparticles; and tailoring proteins for nanoparticle adsorption close to the protein site of interest without losing protein activity.
In conclusion, Narayana notes that there are other areas of research that might benefit from their study. These include screening potential small molecules from drug libraries of various diseases; rational drug design for closely-related targets pertaining to the active site; designing tags for orthogonal imaging without affecting the catalytic activity by binding to unique surface pockets; understanding the specific interactions of drugs based on C-peptide; and in vivo protein imaging.
More information: SERS and MD simulation studies of a kinase inhibitor demonstrate the emergence of a potential drug discovery tool, Proceedings of the National Academy of Sciences, Published online before print on June 27, 2014, doi:10.1073/pnas.1402695111
Journal information: Proceedings of the National Academy of Sciences
© 2014 Medical Xpress
.jpg)