Not only in DNA's hands: Epigenetics has large say in blood formation
Blood stem cells have the potential to turn into any type of blood cell, whether it be the oxygen-carrying red blood cells, or the many types of white blood cells of the immune system that help fight infection. How exactly is the fate of these stem cells regulated? Preliminary findings from research conducted by scientists from the Weizmann Institute and the Hebrew University are starting to reshape the conventional understanding of the way blood stem cell fate decisions are controlled thanks to a new technique for epigenetic analysis they have developed. Understanding epigenetic mechanisms (environmental influences other than genetics) of cell fate could lead to the deciphering of the molecular mechanisms of many diseases, including immunological disorders, anemia, leukemia, and many more. It also lends strong support to findings that environmental factors and lifestyle play a more prominent role in shaping our destiny.
The process of differentiation – in which a stem cell becomes a specialized mature blood cell – is controlled by a cascade of events in which specific genes are turned "on" and "off" in a highly regulated and accurate order. The instructions for this process are contained within the DNA itself in short regulatory sequences. These regulatory regions are normally in a "closed" state masked by special proteins called histones to ensure against unwarranted activation. Therefore, to access and "activate" the instructions, this DNA mask needs to be "opened" by epigenetic modifications of the histones so it can be read by the necessary machinery.
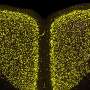
In a paper published in Science, Dr. Ido Amit and David Lara-Astiaso of the Weizmann Institute's Immunology Department, together with Prof. Nir Friedman and Assaf Weiner of the Hebrew University of Jerusalem, charted for the first time histone dynamics during blood development. Thanks to the new technique for epigenetic profiling they developed, in which just a handful of cells – as few as 500 – can be sampled and analyzed accurately, they have identified the exact DNA sequences, as well as the various regulatory proteins, that are involved in regulating the process of blood stem cell fate.
Their research has also yielded unexpected results: As many as 50% of these regulatory sequences are established and opened during intermediate stages of cell development. This means that epigenetics is active at stages in which it had been thought that cell destiny was already set. "This changes our whole understanding of the process of blood stem cell fate decisions," says Lara-Astiaso, "suggesting that the process is more dynamic and flexible than previously thought."
Although this research was conducted on mouse blood stem cells, the scientists believe that the mechanism may hold true for other types of cells. "This research creates a lot of excitement in the field, as it sets the groundwork to study these regulatory elements in humans," says Weiner. Discovering the exact regulatory DNA sequence controlling stem cell fate as well as understanding its mechanism hold promise for the future development of diagnostic tools, personalized medicine, potential therapeutic and nutritional interventions, and perhaps even regenerative medicine, in which committed cells could be reprogrammed to their full stem cell potential.
More information: Science, www.sciencemag.org/content/ear … science.1256271.full