Credit: 2014, Mary Ann Liebert, Inc., publishers
Highly valuable for modeling human diseases and discovering novel drugs and cell-based therapies, induced pluripotent stem cells (iPSCs) are created by reprogramming an adult cell from a patient to obtain patient-specific stem cells. Due to genetic variation, however, iPSCs may differ from a patient's diseased cells, and researchers are now applying new and emerging genomic editing tools to human disease modeling, as described in a comprehensive Review article published in Stem Cells and Development.
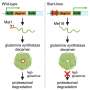
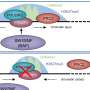
In "Genomic Editing Tools to Model Human Diseases with Isogenic Pluripotent Stem Cells," Ihor Lemischka, Huen Suk Kim, Jeffrey Bernitz, and Dung-Fang Lee, Icahn School of Medicine at Mount Sinai (New York, NY), provide a detailed overview of the development of patient-specific iPSCs for modeling a disease. The authors describe the many factors that need to be considered when generating an iPSC-based disease model comprised of cells that are genetically identical, and they discuss the advantages and limitations of the three leading genomic editing tools: zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the most recent, the clustered regularly interspaced short palindromic repeat (CRISPR) system.
"As our appreciation of iPSCs as primarily therapeutic screens and disease models matures, we look to advanced gene editing tools to assist in appropriate experimental design. Ihor Lemischka and colleagues provide a much needed examination of the advantages and shortcomings of such techniques," says Editor-in-Chief Graham C. Parker, PhD, The Carman and Ann Adams Department of Pediatrics, Wayne State University School of Medicine, Detroit, MI.
More information: The article is available free on the Stem Cells and Development website until September 30, 2014.
Journal information: Stem Cells and Development
Provided by Mary Ann Liebert, Inc