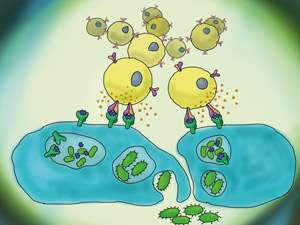
When mucosal-associated invariant T (MAIT) cells (yellow) recognize bacterial products (blue) they promptly release cytokines and exert important immune functions. Credit: Artem Kalinichenko, University Hospital Basel
In an investigation of a specific group of immune cells, researchers at A*STAR identified a limited repertoire of receptors that recognize bacterial targets. This finding led to the further discovery of a previously undetected population of these cells, indicating that they have wider functions than previously thought.
The research team, led by Lucia Mori from the A*STAR Singapore Immunology Network, focused on a population of T cells (which are themselves a type of white blood cell) called mucosal-associated invariant T (MAIT) cells. MAIT cells are abundant in organs such as the gut, liver and kidney, as well as in the blood, and are activated by a wide variety of bacteria.
Like all T cells, MAIT cells express proteins called T-cell receptors (TCRs) that recognize microbes (see image). TCRs are made up of α and β chains that vary to make every TCR unique. Each MAIT cell expresses just one type of TCR. Mori and her team investigated the variety of TCRs expressed by MAIT cells in healthy individuals using a powerful technique called RNA deep sequencing, which enabled more detailed analysis of TCRs than ever before.
The team of Singaporean and Swiss scientists found that the repertoire of TCRs in MAIT cells was extremely limited; this is in contrast with all other types of T cells, which express a much larger variety of TCRs.
"This means that a few original cells that are characterized by a particular TCR β signature clonally expand under stimulation by bacterial products," explains Mori. "These cells were possibly selected as the most efficient at this process, so persist in the body to be continuously restimulated."
Mori and her team also discovered a second population of MAIT cells that had not been previously identified. Similar to the previously described population of MAIT cells, this population has distinct TCR α chains, but the two populations are distributed differently between organs in the body, suggesting that they have different roles.
"MAIT cells monitor intestinal microbial flora and are the best candidates for detecting changes in the gut microbiome induced by pathological changes," says Mori. "Their presence in other organs opens up intriguing areas of research."
The group now hopes to build on this work and gain further insight into the functions of MAIT cells.
"We are investigating a large set of bacteria for their capacity to stimulate all MAIT cell populations and are looking for additional microbial products that can stimulate these cells," says Mori.
More information: Lepore, M., Kalinichenko, A., Colone, A., Paleja, B., Singhal, A et al. "Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRβ repertoire." Nature Communications 5, 3866 (2014). dx.doi.org/10.1038/ncomms4866
Journal information: Nature Communications