Study uncovers secrets of a clump-dissolving protein

Workhorse molecules called heat-shock proteins contribute to refolding proteins that were once misfolded and clumped, causing such disorders as Parkinson's disease, amyotrophic lateral sclerosis, and Alzheimer's disease. James Shorter, PhD, an associate professor of Biochemistry and Biophysics, at the Perelman School of Medicine at the University of Pennsylvania, has been developing ways to "reprogram" one such protein - a yeast protein called Hsp104—to improve its therapeutic properties.
But precise knowledge about the mechanisms by which Hsp104 works to fix misshapened and clumped proteins has been lacking. Now, Shorter and his colleagues have discovered that a previously disregarded part of the Hsp104 structure, the N-terminal domain (NTD), located at one end of the Hsp104 molecule, is a major player in its protein-busting powers. Their work was published in Molecular Cell.
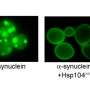
"We've defined in unprecedented detail the mechanism by which Hsp104 dissolves its natural substrate, Sup35 prions," says Shorter. "We found that the N-terminal domain of Hsp104 allows the enzyme to function in a way that enables the disintegration of the prion." Prions are "infectious" proteins that cause disease in humans, but can be beneficial in yeast.
While Hsp104 is found in the vast majority of less complex organisms on the planet, it was somehow lost in the evolution of lower forms of life to more complex animals and humans. "It's baffling in many ways," says Shorter. "We don't understand quite why Hsp104 was lost. But it could be useful in a therapeutic setting because we could add back an activity that humans don't really have: the ability to rapidly dissolve and refold prions." Previously, Shorter and colleagues defined a set of human heat shock proteins that can slowly dissolve prions.
Although previous work by Shorter and others had shown that the middle section of Hsp104 was vital for its clump-busting activity, the N-terminal domain was thought to be relatively unimportant. "Researchers had thought it was a more dispensable domain," says lead author, Elizabeth Sweeny, PhD, a former graduate student in the Shorter lab who is now a postdoctoral fellow at the Massachusetts Institute of Technology. "We reveal in this paper that when you give Hsp104 a very difficult protein clump to break up, like those seen in neurodegenerative disease protein inclusions, it actually becomes very important."
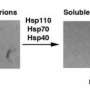
Shorter and his collaborators used small-angle X-ray scattering (SAXS) to examine the role of the Hsp104 N-terminal domain by deleting it from the enzyme and testing it under different conditions. When Hsp104 lacking the NTD (Hsp104?N) is introduced into the formation of the Sup35 prions in a test tube, it promoted prion formation, instead of solubilizing prions. The researchers also observed that, while Hsp104 attacks the Sup35 prion by breaking up the head and tail contacts that hold the prion together, Hsp104?N was unable to do likewise. Hsp104?N is able to dissolve disordered protein aggregates but cannot break down prions due to their increased stability.
Sweeny found that Hsp104 - normally shaped like a short, hexagonal tube—works like a peristaltic pump that shuttles molecules through its central channel. ATP, the cell's energy molecule, is the fuel that powers the pump.
The altered structure of Hsp104?N greatly impairs this normal mechanism, affecting its ability to break apart Sup35 and other prions. Sweeny notes, "Hsp104 extracts individual proteins from the prion fibril by pumping them through its central channel and that's how it dissolves them. The N-terminal domain of Hsp104 allows the enzyme to function in a more powerful way that enables dissolution of the very stable Sup35 yeast prion."
These new insights into the workings of Hsp104 open up a range of new research directions: "We've advanced to a new level of understanding that will help us design and engineer the enzyme to work better against the human proteins that are causing issues in disease," Shorter notes. "Our next step will be to achieve even greater insight into Hsp104 structure. Our level of structural resolution is only about 7 or 8 angstroms at the moment and we'd like to get that down to the atomic level. Another goal is to engineer the N-terminal domain to work better against clumped human disease proteins. Prior to this paper, we wouldn't even have thought about doing that because the domain was not considered important. That's one of the key findings from this study."
Journal information: Molecular Cell
Provided by University of Pennsylvania School of Medicine