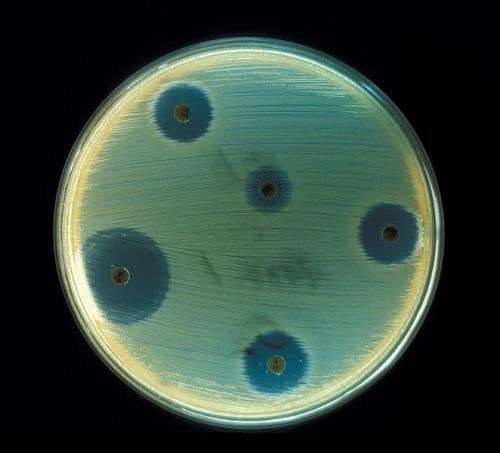
Staphylococcus aureus - Antibiotics Test plate. Credit: CDC
Scientists at the University of Birmingham have identified a new mechanism of antibiotic resistance in bacterial cells which could help us in understanding, and developing solutions to, the growing problem of antibiotic resistance.
The research, published today in PNAS, describes the way in which a strain of Salmonella within a patient was able to develop resistance to the commonly used antibiotic drug ciprofloxacin.
The patient in question was admitted to hospital for repair of a leaking abdominal aortic aneurysm graft, and was treated for a disseminated Salmonella infection.
Through isolates taken over the course of 20 weeks, the team used whole genome sequencing to reveal a mutation in the bacterial cells that allowed them to become resistant to the effects of some antibiotics.
Dr Jessica Blair explained, "We cannot know for sure when this mutation happened within this strain. What we do know is that it developed soon after this patient was given ciprofloxacin to treat the infection. It's further evidence that, when it comes to the issue of antibiotic resistance, we are coming up against a very capable and complex adversary."
Bacteria can become resistant to antibiotics in several different ways. One way is through efflux pumps, bacterial vacuum cleaners, which pump antibiotics from inside bacterial cells to the outside where they are unable to have any effect.
The previously unobserved mutation found in the bacteria isolated from this patient alters the efflux pump. The researchers showed that the mutation makes it more efficient at pumping some antibiotics, including ciprofloxacin, out of the bacterial cells.
Laura Piddock, Professor of Microbiology at the University of Birmingham, BSAC Chair in Public Engagement and Director of Antibiotic Action said, "We have long advocated that the issue of antibiotic resistance was, in the words of Dame Sally Davies, Chief Medical Officer "a ticking time bomb" and that urgent action was needed to stem resistance and identify solutions to the near empty antibiotic pipeline."
"Our study further highlights the need for increased understanding about antibiotic resistance, not least to inform future strategies to both minimise and prevent antibiotic-resistant bacteria arising when new treatments become available."
The team are hopeful that such insights into the mechanisms by which bacteria become resistant to antibiotics will help to design smarter therapies and drugs.
In this instance, the treatments would be designed to avoid the impact of the particular mutation. This is a realistic aspiration, as surprisingly the team also found that some antimicrobial compounds were pumped out poorly by the mutated pump and so had enhanced antibiotic activity.
Antibiotic resistance is becoming the subject of increased focus through the actions of such groups as Antibiotic Action. The World Health Organisation recently warned that "many common infections will no longer have a cure and, once again, could kill unabated."
Professor Piddock added, "Antibiotic use and resistance is still increasing, but it's not surprising with the widespread and often indiscriminate use of these invaluable medicines. Though we don't want to be seen as scaremongering, we've long passed the point at which we can turn a blind eye to the growing threat."
More information: AcrB drug-binding pocket substitution confers clinically relevant resistance and altered substrate specificity, PNAS, www.pnas.org/cgi/doi/10.1073/pnas.1419939112
Journal information: Proceedings of the National Academy of Sciences
Provided by University of Birmingham