In Africa, a deadly salmonella strain takes hold
Salmonella is an infectious agent with many faces, appearing in a multitude of strains affecting animals and humans. A distinct form of the bacterial invader has emerged in sub-Saharan Africa and is responsible for severe epidemic outbreaks. Its unusual characteristics—including a high rate of lethality, invasiveness, atypical symptomatolgy and resistance to multiple antibiotics—are of rising concern.
In a new study, Cheryl Nickerson and her colleagues at the Biodesign Institute at Arizona State University and NASA Johnson Space Center demonstrate for the first time that this pathogen can cause lethal infections not only in humans but in mice, a finding which could potentially extend to other hosts as well.
The salmonella strain used in this study, D23580, belongs to a group of closely related strains collectively known as ST313, and was shown to more rapidly reach and colonize tissues of the spleen and gallbladder in mice, compared with a well-characterized "classic" salmonella strain.
In results appearing in the journal PLOS Neglected Tropical Diseases, lead authors Jiseon Yang and Jennifer Barrila also establish a critical variable of the pathogen known as LD50—a measure of the median lethal dose (LD) necessary to produce a fatal infection—marking the first report of the entire natural course of disease for any ST313 strain.
Developing effective means to diagnose and treat deadly salmonella infections, including those caused by ST313, will require a more thorough understanding of the strategies used by such pathogens to infect the body. Establishing LD50 is a necessary step for examining the trajectory of salmonella infection and developing effective vaccines and therapies to combat it.
"Despite being one of the best characterized pathogens, we still have limited knowledge of the mechanisms used by salmonella to cause disease in humans, including the multidrug-resistant ST313 isolates associated with rampant atypical disease and high mortality in sub-Saharan Africa," said Nickerson, who is also a professor of microbiology at ASU's School of Life Sciences.
The current study offers new insight into the virulence and pathogenesis properties of model ST313 strain, D23580, which shows both key similarities and differences between classic Typhimurium and Typhi strains in its virulence and pathogenesis-related properties, thus offering clues as to how it may cause disease in humans.
Unwelcome visitor
The rod-shaped bacterium salmonella infects a broad range of warm- and cold-blooded animals around the world. The pervasiveness of the pathogen owes much to its aptitude for infection under varying conditions and its stubborn durability—it can survive freezing temperatures as well as the highly acidic conditions found in the human stomach and the low oxygen and detergent-like bile stresses present in the intestine.
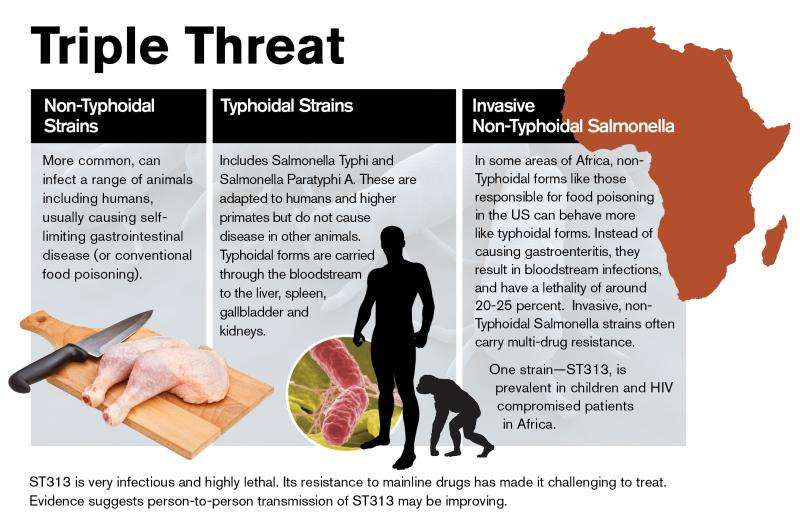
Two broad categories of salmonella—known as typhoidal and non-typhoidal—menace human populations. Strains of the former pathogen include salmonella Typhi and Paratyphi and are responsible for typhoid fever. They are so-called host-restricted pathogens, targeting only humans and higher primates.
Symptoms of typhoid fever range from moderate to severe. Infection can lead to serious complications, as the bacteria are carried systemically through the lymphatic system of the intestine to other organs including the liver, kidneys, gallbladder and spleen. Typhi flourishes under conditions of poor hygiene and is typically a problem in underdeveloped countries, where it is acquired by ingesting food or water contaminated with feces from an infected person. A vaccine for typhoid offers protection in around 50-70 percent of cases. The condition, if not successfully treated, can lead to shock, organ failure and death.
By contrast, non-typhoidal forms of salmonella are responsible for foodborne disease, sickening tens of millions worldwide each year in both developed and underdeveloped countries, according to the World Health Organization. In healthy individuals, the infection is self-limiting and restricted to the intestinal tract. There are more than 2,500 different variants of non-typhoidal salmonella.
Unlike its typhoidal brethren, non-typhoidal salmonella strains are zoonotic—infecting a wide variety of animal hosts. Usually, non-typhoidal salmonella are transmitted through the consumption of contaminated food, with meat, poultry, eggs and milk among the primary culprits. Currently, there is no effective vaccine to protect against non-typhoidal salmonella food poisoning.
The onset of salmonella-induced foodborne illness usually occurs within 12-72 hours of infection and is characterized by fever, abdominal pain, diarrhea, nausea and vomiting, typically lasting four to seven days. Although most people make a full recovery, foodborne illness poses serious risk to the very young, elderly and other immune-compromised populations.
Agent of destruction
The current study focuses on ST313, a salmonella variant that has rapidly emerged in sub-Saharan Africa. It is a major cause of fatal bacterial infections in young children affected by malaria, severe anemia and/or malnutrition as well as adults infected with HIV. Infection with ST313 can also lead to septicemia and meningitis.
While technically categorized as S. Typhimurium, a primary non-typhoidal salmonella, ST313 nevertheless bears some resemblance to typhoid strains of the bacterium. Earlier studies seem to suggest the variant is evolving to become more host-restricted to humans, compared with better-known non-typhoidal strains.
Notably, ST313 often does not produce symptoms of gastroenteritis typically associated with foodborne illness. Rather, invasive non-typhoidal salmonella (iNTS) strains like ST313 are a leading cause of bloodstream infections in sub-Saharan Africa. Such infections are often fatal.
In addition to causing high mortality, iNTS infections display resistance to multiple antibiotics, making effective treatment challenging, particularly in impoverished regions lacking access to effective alternatives. Fatality rates for children range from 20-25 percent, while rates for HIV-positive patients may run as high as 50 percent.
An elusive peril
Thus far, no animal reservoir has been identified for ST313. Unlike conventional foodborne NTS infections, the transmission route for ST313 appears to be human-to-human. Genetic studies of iNTS strains obtained from Malawi, where ST313 is highly prevalent; indicate the strain may be losing genetic diversity, becoming a more specialized pathogen, similar to S. Typhi.
The current study, however, shows that ST313 strain D23580 could also infect mice and thus retains characteristics associated with classic NTS infection. However, biochemical and phenotypic assays indicated that D23580 also exhibits important differences between classic NTS and typhoidal strains. Collectively, these results provide further evidence that this emerging pathogen is distinct from classic salmonella strains.
Although earlier research showed that ST313 is capable of causing systemic infections in chickens and mice, it was unclear whether it could produce lethal infections in non-human animals. Using a mouse model, the current study demonstrates that indeed it can.
Compared with common strains of NTS, the ST313 strain more easily colonized the spleen and gallbladder in mice. Alarmingly, measurement of the LD50 of this strain revealed that it is more than four times more virulent in a mouse model compared with a previously tested "classic" S. Typhimurium strain.
The authors also showed that the swimming ability of the ST313 strain was enhanced, compared with its closely related salmonella counterparts. In vitro studies confirmed the ability of the ST313 strain to better tolerate low pH conditions of the stomach—another key factor affecting the duration and severity of ST313 infections compared with other NTS strains. The authors suggest that heightened acid resistance in ST313 strains may facilitate person-to-person transmission of the bacterium.
The current study marks the first time that the entire disease course for an ST313 strain was tracked, establishing the pathogen's increased capacity for infection, with lethal results. The challenges for accurate diagnosis and treatment of those stricken with ST313 and similar invasive non-typhoidal variants are acute.
"The morbidity and mortality associated with these multidrug-resistant, non-typhoidal salmonella strains continues to be a major public health concern, as does the potential risk for global spread of this infectious disease," Yang said. "Our findings in this study provide an important laboratory benchmark that researchers worldwide can use for their investigations into the disease-causing mechanisms of these deadly pathogens."
More information: "Characterization of the Invasive, Multidrug Resistant Non-typhoidal Salmonella Strain D23580 in a Murine Model of Infection." PLoS Negl Trop Dis. 2015 Jun; 9(6): e0003839. doi: 10.1371/journal.pntd.0003839