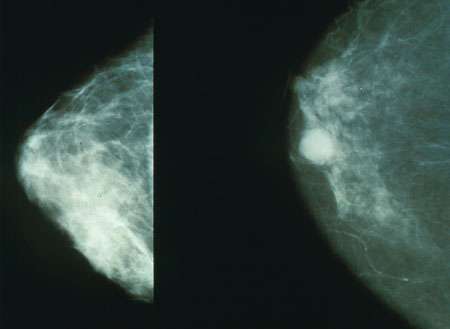
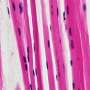
Mammograms showing a normal breast (left) and a breast with cancer (right). Credit: Public Domain
Running large, multi-gene sequencing panels to assess cancer risk is a growing trend in medicine as the price of the technology declines and more precise approaches to cancer care gain steam. The tests are particularly common among breast and ovarian cancer patients. However, questions remain about the growing list of mutations and their suspected, but unproven association with breast and ovarian cancer risk.
In a new study published in the American Journal of Human Genetics, a team led by researchers at the University of Pennsylvania's Abramson Cancer Center (ACC) found that expanding the panel beyond the known breast/ovarian cancer-specific genes in these patients does not add any clinical benefit. Instead, testing for additional cancer susceptibility genes produced more questions than answers for breast and ovarian cancer patients.
Genetic panels sometimes reveal mutations in genes that are associated with an increased risk in developing cancer. BRCA 1 and BRCA 2 genes are prime examples, and those results may prompt women to opt for mastectomies and ovary removal surgery, which greatly decrease their risk of developing those cancers. However, there is not yet guidance for clinicians on how to care for patients who exhibit other types of mutations, such as those in CHEK2 and ATM .
To determine if there was a benefit to expanding the gene panel to include non-breast/ovarian cancer and even non-cancer susceptibility genes, the team, led by Katherine Nathanson, MD, a professor of Translational Medicine and Human Genetics, associate director of Population Science and Chief Oncogenomics Physician for the ACC, performed whole exome sequencing and then analyzed an 180 gene panel (including 25 breast/ovarian cancer specific-susceptibility genes, 123 other cancer-susceptibility genes, and 32 genes related to cardiovascular disease risk) in 404 individuals in 253 families with breast and/or ovarian cancer.
This project was done in collaboration with researchers from the Mayo Clinic, Memorial Sloan Kettering Cancer Center and City of Hope as part of the SIMPLEXO (Simplifying Complex Exomes) consortium.
The study also evaluated the clinical utility of a variant classification methodology based on the American College of Medical Genetics and Genomics (ACMG) guidelines to help define mutations. These guidelines were recently published by the ACMG partly to address the inconsistencies in the classification of mutations among clinical laboratories. However, a "real world" methodology using the guidelines in cancer susceptibility had not been well studied or reported.
The analysis underscores the complexities of genetic testing and counseling, which only continues to increase in today's world. Additionally, genetic testing identifies far more mutations, some referred to as "variants of uncertain significance," or VUS, that clinicians know very little about or how to act on.
Using the ACMG guidelines, the team identified 1,605 variants, and found that 11 percent of the patients without BRCA 1/2 mutations had clinically actionable mutations in other cancer susceptibility genes. Most confer a moderate increased risk of breast or ovarian cancer; only two additional families had mutations in non-breast cancer susceptibility genes (ATR and MSH6).
Further, 12 percent of patients had VUSs in the well-established breast cancer susceptibility genes. When the non-breast/ovarian cancer susceptibility genes were included as well, however, the VUS rate jumped significantly: 78 percent of families had variants with no known clinical significance.
"Adding on the additional cancer susceptibility genes to the 'breast cancer susceptibility' genes opened up more questions than it answered," Nathanson said. "This study therefore adds to prior findings of ours, and demonstrates little incremental utility to testing non-breast cancer susceptibility genes in breast cancer families."
The team also looked at the rate of mutations and VUSs in genes associated with cardiovascular disease risk, which are recommended for return of results by the ACMG as "secondary findings." They found very few families with clinically actionable mutations. They also found some carriers of autosomal recessive cancer susceptibility gene mutations. Adding all the genes together, 95 percent of patients had at least one VUS or mutation identified, and nearly 30 percent of patients had five or more VUSs.
"These findings add layers of complexity in counseling for cancer risk," said the study's first author Kara Maxwell, MD, PhD, an instructor in the division of Hematology/Oncology. "A patient would now be expected to understand not only that a genetic finding could be found that may or may not be related to the cancer in their family, but that genetic variants could be found that may or may not put themselves and their family members at risk for a disease they weren't even thinking about at the time."
According to the researchers, the methodology they developed to apply the ACMG guidelines was of clinical utility and worked well to classify known variants.
Journal information: American Journal of Human Genetics
Provided by University of Pennsylvania School of Medicine