Penn software helps to identify course of cancer metastasis, tumor 'evolution'
Individual cells within a tumor are not all the same. This may sound like a modern medical truism, but it wasn't very long ago that oncologists assumed that taking a single biopsy from a patient's tumor would be an accurate reflection of the physiological and genetic make-up of the entire mass.
Researchers have come to realize that cancer is a disease driven by the same "survival of the fitter" forces that Darwin proposed drove the evolution of life on Earth. In the case of tumors, however, individual cells are constantly evolving as a tumor's stage advances. Mobile cancer cells causing metastasis are a deadly outcome of this process.
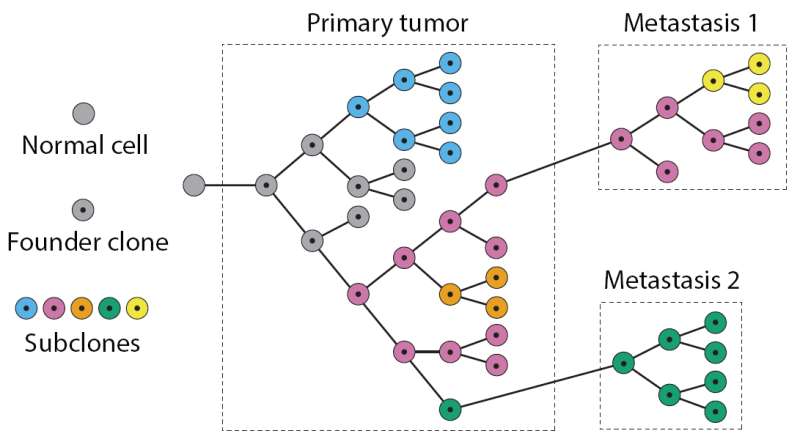
Tumors also differ among patients with the same type of cancer, so how is a physician able to prescribe a tailored regimen for the patient? To start to address this conundrum, an interdisciplinary team from the Perelman School of Medicine and the Wharton School at the University of Pennsylvania developed Canopy, an approach to infer the evolutionary track of tumor cells by surveying two types of mutations - somatic copy number alterations and single-nucleotide alterations - derived from multiple samples taken from a single patient. They demonstrated the approach on samples from leukemia and ovarian cancer, along with samples from a human breast cancer cell line. Overall, the evolution of a tumor involves the accumulation of mutations of all types collectively influencing the fitness of tumor cells.
The team, Yuchao Jiang, a doctoral student in the Genomics and Computational Biology program, Yu Qiu, PhD, a postdoctoral researcher in the lab of coauthor Andy Minn, MD, PhD, an assistant professor of Radiation Oncology, and Nancy R Zhang, PhD, an associate professor of Statistics in the Wharton School, published their findings online in the early edition of the Proceedings of the National Academy of Sciences.
"The make-up of a tumor for a given patient is often a mixture of multiple distinct cell populations that differ in genetic make-up, gene expression, and physiology," Jiang said. "This heterogeneity contributes to failures of targeted therapies and to drug resistance based on old thinking that tumors are homogenous masses."
However, Canopy takes these differences into account because it uses data from multiple slices of the same tumor in space and time, as opposed to sampling in one spot at one time point, the way it is currently done in most sequencing studies. Canopy is an open-source software so oncologists will be able to use it to identify potential biomarkers for different cancer cell populations within tumor specimens that are associated with drug resistance and invasive malignancy, among other characteristics.
"Data input for Canopy from the clinic would come from tissue samples from the same patient - from normal tissue and from different places on the tumor and at different times," Jiang said. "Canopy can then be used to infer the evolutionary history of the tumor to better understand how the tumor evolved and to find potential biomarkers." The goal for Canopy is to help new patients at an early stage, using biomarkers to ascertain a correct diagnosis and prognosis.
The team also used animal models to refine their understanding of a metastasis model for breast cancer. After injecting human breast cancer cells into mice they observed where the primary breast cancer cells metastasized—lung, bone, or other tissues. The site-specific metastatic samples, as well as the primary cancer cell lines were sequenced by whole-exome sequencing, from which somatic single-nucleotide alterations and copy number mutations were profiled. The relationships reconstructed by Canopy showed that, compared to the primary breast cancer cell lines, the metastatic samples from different organs had distinct genotypic patterns. Canopy identified metastasis-site-specific mutations for this specific malignant breast cancer cell line. The concept behind this proof-of-concept study can be applied to larger studies with the goal of identifying genomic changes that serve as prognostic markers for the development of distant metastasis.
By assigning mutation types to different parts of a tumor's "evolutionary tree," oncologists will be able to predict the site and severity of metastasis, and potentially, better courses of treatment. Applying Canopy to a larger cohort of samples is one of the team's future directions.
In addition, says Zhang, recent advances in single-cell sequencing make possible the study of tumors at the single-cell level. However, reliable simultaneous profiling of copy number and single nucleotide mutations by single-cell sequencing is still in its infancy. "Our work shows that traditional bulk sequencing can lead to accurate tumor subclone identification, if the researcher is willing to sequence multiple slices of the tissue," she said. "Bulk tissue sequencing can play an important part in our understanding of tumor heterogeneity, and in the coming years, experimental designs that combine bulk tissue sampling and single cell analysis need to be better explored."
More information: Yuchao Jiang et al. Assessing intratumor heterogeneity and tracking longitudinal and spatial clonal evolutionary history by next-generation sequencing, Proceedings of the National Academy of Sciences (2016). DOI: 10.1073/pnas.1522203113