December 8, 2016 report
Researchers find DNA mutation that led to change in function of gene in humans that sparked larger neocortex
(Medical Xpress)—A team of researchers at the Max Planck Institute has found what they believe is the DNA mutation that led to a change in function of a gene in humans that sparked the growth of a larger neocortex. In their paper published in the journal Science Advances, the team describes how they engineered a gene found only in humans, Denisovans and Neanderthals to look like a precursor to reveal its neuroproliferative effect.
A year ago, another team of researchers found the human gene that most in the field believe was a major factor in allowing the human brain to grow bigger, allowing for more complex processing. In this new effort, the researchers have found what they believe was the DNA change that arose in that gene.
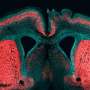
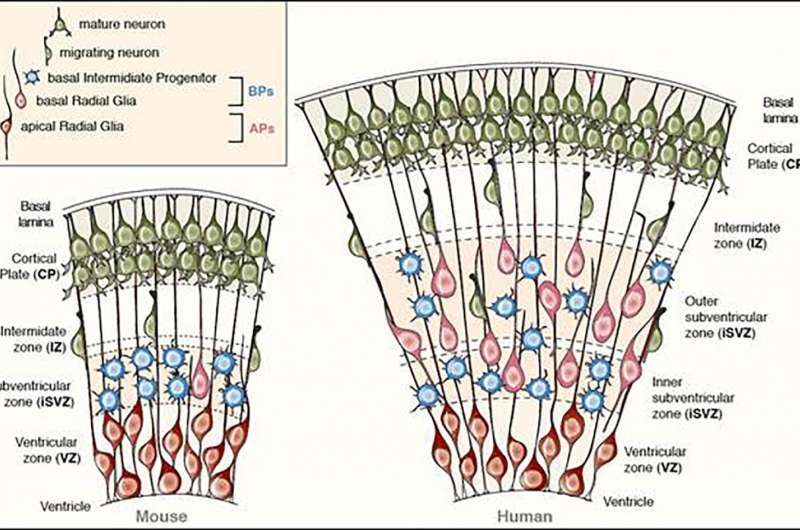
To pinpoint that change, the researchers engineered the unique ARHGAP11B gene to make it more similar to the ARHGAP11A gene, which researchers believe was a predecessor gene—they swapped a single nucleotide (out of 55 possibilities) for another and in so doing, found the ARHGAP11B gene lost its neuroproliferative abilities. This, the team claims, shows that it was a single mutation that allowed humans to grow bigger brains. Such a mutation, they note, was not likely due to natural selection, but was more likely a simple mistake that occurred as a brain cell was splitting. Because it conferred an advantage (the ability to grow higher than normal amounts of brain cells) the mutation was retained through subsequent generations. They also point out that such a mutation would have resulted specifically in a larger neocortex—a portion of the cortex that has been associated with hearing and sight. Prior research has also found that this region of the brain is likely the part of the brain that has most recently evolved.
The researchers also note that their research showed the mutation occurring just 1 million years after the human line split from chimpanzees—which was approximately 5 to 6 million years ago. Since that time, other research has shown the human brain has experienced several growth spurts leading to advances in intelligence and the ability to reason.
More information: M. Florio et al. A single splice site mutation in human-specific ARHGAP11B causes basal progenitor amplification, Science Advances (2016). DOI: 10.1126/sciadv.1601941
Abstract
The gene ARHGAP11B promotes basal progenitor amplification and is implicated in neocortex expansion. It arose on the human evolutionary lineage by partial duplication of ARHGAP11A, which encodes a Rho guanosine triphosphatase–activating protein (RhoGAP). However, a lack of 55 nucleotides in ARHGAP11B mRNA leads to loss of RhoGAP activity by GAP domain truncation and addition of a human-specific carboxy-terminal amino acid sequence. We show that these 55 nucleotides are deleted by mRNA splicing due to a single C→G substitution that creates a novel splice donor site. We reconstructed an ancestral ARHGAP11B complementary DNA without this substitution. Ancestral ARHGAP11B exhibits RhoGAP activity but has no ability to increase basal progenitors during neocortex development. Hence, a single nucleotide substitution underlies the specific properties of ARHGAP11B that likely contributed to the evolutionary expansion of the human neocortex.
© 2016 Medical Xpress