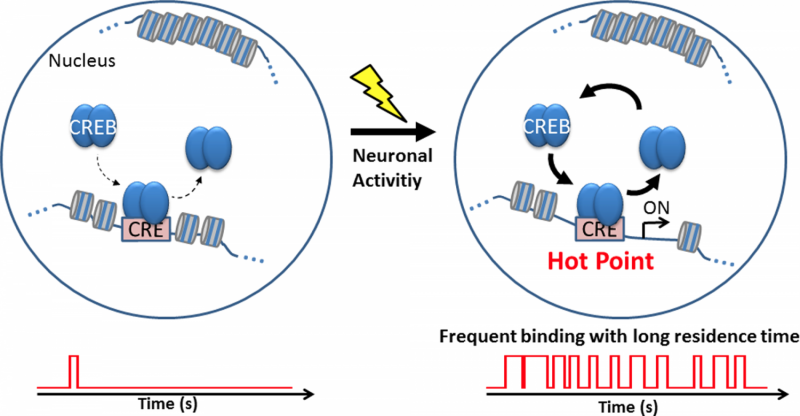
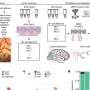
A model of activity-dependent transcription mediated by CREB in cortical neurons. A model of the frequent interaction of CREB with CRE to promote transcription initiation in response to neuronal activity. Credit: Osaka University
Neuronal activity mediates the formation of neuronal circuits in the cerebral cortex. These processes are regulated by the transcription factor CREB, which regulates gene expression in neuronal activity-dependent processes. Neuronal activity enhances CREB-mediated transcription but the mechanisms remain unclear.
CREB binds to a cAMP response element (CRE) in the promoter region of its target genes. Assembly and disassembly of CREB-CRE interactions control spatiotemporal gene expression in the nucleus. However, how CREB interacts with CRE in activity-dependent mechanisms is not known.
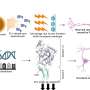
In a new study from Osaka University, led by Nobuhiko Yamamoto, Hironobu Kitagawa, Noriyuki Sugo and colleagues investigated how neuronal activity influences CREB dynamics. Fluorescent-tagged CREB constructs were expressed in primary cortical neurons and visualized by staining fixed cells. Single CREB molecules were predominantly detected in the nucleus and some were observed at sites where gene transcription was taking place.
The movement of single CREB molecules in living cortical neurons was monitored by real-time imaging. Interestingly, some CREB molecules disappeared almost immediately while a fraction remained in the same location for a longer duration. Using mutant forms of CREB that cannot bind to CRE, the researchers showed that these CREB molecules were interacting with CRE.
To investigate how neuronal activity affects CREB-CRE interactions and subsequent gene transcription, the authors treated cortical neurons with pharmacological agents that alter neuronal activity. Neuronal activity did not affect the time that CREB resided in the nucleus.
Next, the research team investigated whether neuronal activity regulates the spatial distribution of CREB in the nucleus. They stimulated cortical neurons and monitored CREB localization by real-time imaging. Interestingly, the number of locations where CREB resided for long periods increased considerably after neuronal activation. These findings indicated that neuronal activity increases CREB-CRE interactions in the nucleus.
The findings provide novel insights into the regulation of gene expression by neuronal activity. Based on the results of this study, neuronal activity may contribute to CREB-dependent gene expression by increasing the binding of CREB to specific genomic sites.
More information: Hironobu Kitagawa et al. Activity-Dependent Dynamics of the Transcription Factor CREB in Cortical Neurons Revealed by Single-Molecule Imaging, The Journal of Neuroscience (2016). DOI: 10.1523/JNEUROSCI.0943-16.2016
Journal information: Journal of Neuroscience
Provided by Osaka University