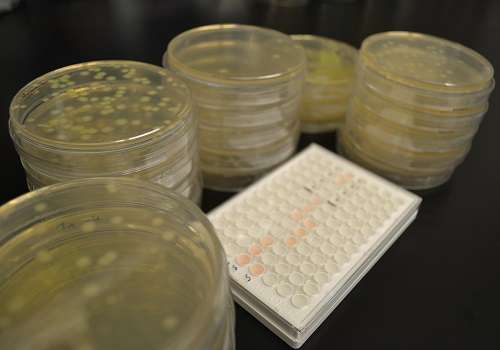
Researchers at KAUST have uncovered a gene circulating in the Jeddah wastewater networks that confers resistance to last-resort antibiotics. The team also showed that a gene is carried by a strongly resistant strain of E.coli, PI7, which can survive for long distances through the municipal wastewater system. Credit: Pei-Ying Hong
An antibiotic resistant strain of bacteria found circulating in Jeddah's municipal wastewater could have severe implications for public health.
The dramatic rise in bacterial superbugs that are resistant to last-resort antibiotics poses a significant global health threat. Recently, KAUST researchers uncovered incidences of a resistance-conferring gene carried by bacteria in municipal wastewater in Jeddah, Saudi Arabia.
The recently discovered New Delhi metallo beta-lactamase enzyme (NDM), which is carried by the blaNDM-1 gene, confers resistance to last-resort antibiotics called carbapenems. Without carbapenems, our ability to tackle potentially lethal bacterial infections is greatly reduced. The situation is exacerbated by people travelling widely, enabling resistant bacteria to spread further and faster than ever before.
"There has been an increase in infections caused by NDM-positive bacteria reported in hospitals across the Gulf," explained Assistant Professor of Environmental Science and Engineering and member of the University's Water Desalination and Reuse Center Pei-Ying Hong, who led the project team. "We began to wonder if viable NDM-positive pathogens were circulating in local communities via presence in Jeddah's municipal wastewater networks. We were also interested to see if pathogen levels spiked during times of religious pilgrimage, a time when millions of people come to Saudi Arabia from across the world."
One problem with the current infrastructure in Jeddah is that only around 50 percent of wastewater is treated in centralized sewage plants. The remainder is partially treated in septic tanks and later discharged into the environment.
Hong's team used the state-of-the art equipment in the University's Bioscience Core Lab to analyze wastewater from a Jeddah treatment plant over the course of a year. Using a molecular technique, they found that up to 104 copies of blaNDM-1 were present in every cubic meter of wastewater. While the abundance of the gene fluctuated throughout the year, there was no spike during the Hajj pilgrimage, suggesting that blaNDM-1 was already locally prevalent in bacterial communities.
When the team identified that the gene was carried by a strain of Escherichia coli called PI7, they conducted genomic investigations into E. coli PI7 to reveal concerning results.
"E.coli PI7 possesses a resistance profile organized in a genetic structure that is similar to one discovered in another bacterium in Taiwan," said the study's first author Ph.D. student David Mantilla-Calderon. "This highlights the global mobility of this genetic structure. Not only this, but E. coli PI7 is resistant to a wide spectrum of antibiotics and its genetic make-up encourages gene transfer to other bacteria." The results also indicate that E. coli PI7 remains viable even after passing through sewage networks over long distances. This raises questions about its potential environmental persistence.
Hong told us that further investigations into the persistence of blaNDM-1 in the environment are vital. She also stressed the importance of improving wastewater treatment infrastructures and of having careful surveillance of pathogens in such networks.
More information: David Mantilla-Calderon et al. Isolation and Characterization of NDM-Positive Escherichia coli from Municipal Wastewater in Jeddah, Saudi Arabia, Antimicrobial Agents and Chemotherapy (2016). DOI: 10.1128/AAC.00236-16
Journal information: Antimicrobial Agents and Chemotherapy