Researchers use new computational method to define immune cell interactions
Immunotherapy, harnessing a patient's immune system to help fight cancer, has shown much promise as a potential cancer treatment. In a newly published study, a research team at Dartmouth-Hitchcock's Norris Cotton Cancer Center (NCCC) illustrates that complex interactions between different immune cell types in the tumor microenvironment play meaningful roles in patient survival. The team used a new computational method to infer immune cell infiltration from patient gene expression data, enabling them to quickly calculate the personal immune response profile for thousands of patients. Their findings, Systematic Pan-Cancer Analysis Reveals Immune Cell Interactions in the Tumor Microenvironment will be published in the next issue of Cancer Research.
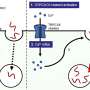
The study, which took place in the Chao Cheng laboratory at NCCC was led by Fred Varn, BS, a graduate student and PhD candidate in the Cheng laboratory. "In our publication, we were curious about how different immune cells affect each other in different cancer types. Some immune cells, such as CD8+ T cells, have tumor-killing capabilities while others, such as T-regulatory cells and certain myeloid cells, can suppress these attacks. Understanding how these cell types infiltrate different tumors and the effect these cells have on each other and the patient can help us understand how to better harness the power of the immune system for cancer therapy" said Varn.
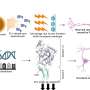
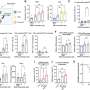
The team used a computational method to outline the patterns of immune infiltration in different tumor types. They found that most immune cell types tend to co-infiltrate the tumor together, demonstrating the importance of accounting for the full patient immune response profile when trying to determine the effect of a single immune cell on patient outcome. The study later validates this idea by showing that for several tumor types, the beneficial patient survival effects of "good" (tumor-killing) immune infiltration can be modulated by infiltration from immunosuppressive cell types.
The study additionally examines what causes patients to have different immune infiltration patterns and finds that tumor types with a higher average mutation count tend to have higher immune infiltration. However, when looking within a single cancer type, mutation count was not associated with increased mutation burden, suggesting that other factors may drive the immune infiltration differences between patients with the same type of tumor.
"Our study uses a computational method that can be cheaply and easily applied to patient gene expression profiles to explore patients' baseline tumor immune response" explains Varn. "This information can eventually be used to help identify patients likely to respond to certain immunotherapeutic approaches, as baseline immune infiltration of certain cell types has been implicated as a predictor of response to numerous immunotherapeutic approaches."
Looking ahead, the team hopes to use their method to predict which patients are likely to respond to immunotherapy. To accomplish this, they will need patient datasets that include both the gene expression information that their method uses to infer a patient's immune infiltration profile, and immunotherapy response information. Currently, very few of these datasets have been made publicly available. However, as immunotherapeutic approaches continue to develop, Varn and his team anticipate more of these datasets will be made available, allowing them to eventually use their method to predict immunotherapy responders. "Our study is the first, to our knowledge, that uses computational approaches to examine the effect different immune cells have on each other in the context of the tumor and outline how these interactions affect patient survival" said Varn.
A patient's personal immune response profile can greatly affect their prognosis and can likely affect their response to immunotherapeutic approaches. It is not enough to simply measure the levels of one cell type, as other cell types likely affect immune cell behavior in the tumor. Understanding how to increase tumor-killing cell activity and decrease immunosuppressive cell activity will be an important question to answer going forward.
More information: Frederick S. Varn et al, Systematic Pan-Cancer Analysis Reveals Immune Cell Interactions in the Tumor Microenvironment, Cancer Research (2017). DOI: 10.1158/0008-5472.CAN-16-2490