Study parses influence of genes and environment in metabolic disease
By comparing two strains of mice—one that becomes obese and diabetic on a high-fat diet and another resistant to a high-fat regimen—researchers from the Perelman School of Medicine at the University of Pennsylvania identified genome-wide changes caused by a high-fat diet.
The a team, led by Raymond Soccio, MD, PhD, an assistant professor of Medicine, and Mitchell Lazar, MD, PhD, director the Institute for Diabetes, Obesity, and Metabolism, published their findings online in the Journal of Clinical Investigation (JCI), in addition to an Author's Take video.
"We focused on the epigenome, the part of the genome that doesn't code for proteins but governs gene expression," Lazar said.
Their research suggests that people who may be genetically susceptible to obesity and type 2 diabetes due to low levels of a protein that helps cells burn fat, may benefit from treatments that ultimately increase the fat-burning molecule.
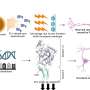
The team looked at the interplay of genes and environment in two types of white fat tissue, subcutaneous fat (under the skin) versus visceral fat around abdominal organs. The latter correlates strongly with metabolic disease. This visceral fat shows major gene expression changes in diet-induced obesity. The JCI study confirmed this relationship—and importantly—extended these findings to show that the epigenome in visceral fat also changes on a high fat diet.
Diet-induced epigenomic changes in fat cells occur at histones - proteins that package and order DNA in the nucleus, which influences gene expression - across the genome. There were also changes in the binding to DNA of an essential fat cell protein, a transcription factor called PPARgamma.
The team next treated obese mice with the drug rosiglitazone, which targets PPARgamma in fat to treat diabetes in people. "While the drug-treated obese mice were more insulin sensitive, we were surprised to see that the drug had little effect on gene expression in visceral fat," Soccio said. "This led us to look at subcutaneous fat and we discovered that this depot is much more responsive to the drug."
"These results are clinically relevant and indicate that the 'bad' metabolic effects of obesity occur in visceral fat, while the 'good' effects of rosiglitazone and other drugs like it occur in subcutaneous fat," Lazar said.
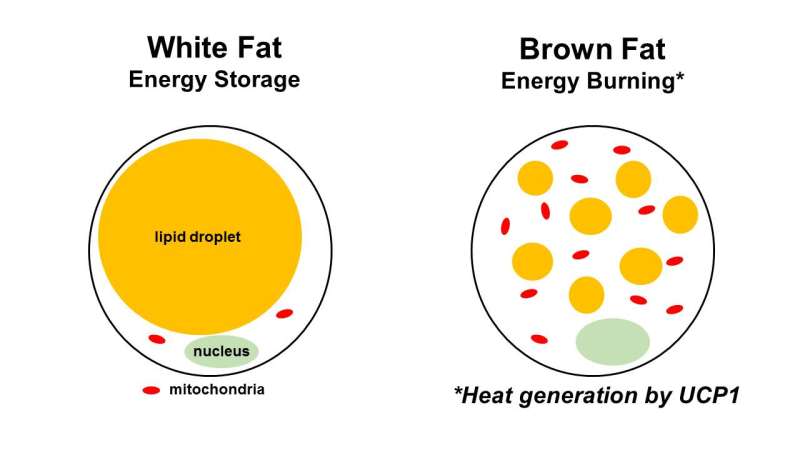
In particular, the drug-induced changes they found in subcutaneous fat reflected the phenomenon of browning, in which white fat takes on characteristics of brown fat, typically in response to cold exposure or certain hormones and drugs.
White fat stores energy, while brown fat dissipates energy by producing heat, mediated by uncoupling protein 1, or UCP1. The most interesting discovery of the study, say the authors, involves UCP1.
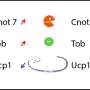
They showed that rosiglitazone, as expected, increases Ucp1 expression in both obesity-prone and obesity-resistant strains of mice. However, in subcutaneous fat of the obesity-resistant mice, Ucp1 expression was high even in the absence of the drug. "But the real surprise came when we looked at the offspring of obesity-resistant and obesity-prone parents, which have one of each parent's version of the Ucp1 gene," Soccio said.
Strikingly, they found that the obesity-prone mouse strain's version of the Ucp1 gene has lower expression and less PPARgamma binding than the obesity-resistant version. This imbalance shows that the obesity-prone mouse strain's Ucp1 is genetically defective, since it is less active than the other strain's version, even when both are present in the same cell nucleus.
In their final experiments, the team asked what happens when browning and Ucp1 expression are activated using rosiglitazone or exposure to cold, both environmental factors. They found that in both cases, total Ucp1 expression goes up as expected, but the obesity-prone strain's defective version of Ucp1 now reaches equal levels to the obesity-resistant strain's version.
"Importantly, we were only changing the mouse's environment with a drug or temperature, not the actual DNA sequence of the Ucp1 gene," Lazar said. "We propose that this result indicates epigenomic rescue of Ucp1 expression in subcutaneous fat cells."
The team is following up the mouse studies using human fat biopsies to figure out the exact DNA sequence differences responsible for variable Ucp1 expression, both in mice and in humans.
The relevance of this study extends even beyond UCP1 and obesity. "Many gene variants are thought to exert their effects by ultimately altering gene expression levels, and this study shows that a genetic predisposition to altered gene expression can be identified and then overcome with treatment," Lazar said. "This is the dream of precision medicine, and hopefully our study is a step in this direction."