Scientists find surprising impact of junk DNA and RNA in cancer
"Human satellite II," an exceptionally high-copy but unexplored sequence of the human genome thought of as "junk DNA," has a surprising ability to impact master regulators of our genome, and it goes awry in 50 percent of tumors, according to a new study published in Cell Reports by scientists at UMass Medical School. When deregulated, these large expanses of repeated nucleotide sequences bind and sequester master regulatory proteins into large nuclear bodies in cancer cells. This likely contributes to epigenetic instability—changes in gene regulation—that are a driving force in the progression of many cancers.
"These satellite repeats are like molecular sponges for master regulatory factors that normally control the expression of other genes," said Jeanne Lawrence, PhD, professor of neurology. "Over time, as cells age and divide, these changes could lead to epigenetic instability with certain cancer-associated genes becoming active."
A giant blank box in the human genome that has yet to be fully sequenced and mapped, HSAT-II consists of millions of copies of a small sequence that is so abundant that it can be seen under a microscope on some chromosomes, particularly chromosome 1. Because HSAT-II DNA is normally methylated (a form of gene regulation), it remains dormant in healthy cells. For this reason, the HSAT-II hasn't been extensively studied and has not been thought to have a function. In fact, standard genomic experiments intentionally screen HSAT-II out of the results.
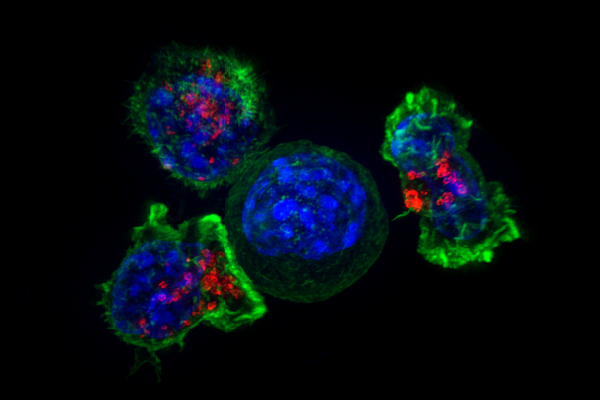
Seeking to understand whether repetitive "junk DNA" might have function, the Lawrence lab used fluorescence in situ hybridization (FISH) to track RNA, and discovered that in half of all tumors studied, HSAT-II was expressed and formed large nuclear accumulations of RNA. To understand why HSAT-II was misregulated in cancer, Dr. Lawrence, Lisa Hall-Anderson, PhD, associate professor of neurology, and Meg Byron, research associate, induced demethylation of HSAT-II DNA, which is common in cancer but not thought to have functional consequence. They discovered that when demethylated, HSAT-II DNA "soaks up" the important regulatory complex known as PRC1 (protein regulator of cytokinesis 1) and, HSATII RNA foci similarly corrals MeCP2 (methyl CpG binding protein 2), resulting in two types of cancer-associated nuclear bodies. These proteins help control a number of other genes responsible for cell differentiation and developmental pathways known to go awry in cancer.
"This repetitive 'junk' in our genome is a giant blank spot where the sequences haven't been mapped. Researchers rarely look at them. This junk, though, may also be what plays a role in control of our genes," said Dr. Hall-Anderson.
Lawrence added, "In cancer biology, genomic instability is a big focus. However, it's not just DNA sequence changes, but regulation that goes awry also. It doesn't matter if the sequence is right if the genes are being misregulated. This is evidence that the epigenome can build on itself and potentially play a big role in cancer formation."
While this work points to a new direction in cancer research, Lawrence and colleagues suggest it will also prompt exploration of what role HSAT-II may play in development of healthy cells.
More information: Lisa L. Hall et al. Demethylated HSATII DNA and HSATII RNA Foci Sequester PRC1 and MeCP2 into Cancer-Specific Nuclear Bodies, Cell Reports (2017). DOI: 10.1016/j.celrep.2017.02.072